Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most fatal malignancies worldwide. Recently, our group identified purine-rich element binding protein alpha (PURα), a single-stranded DNA/RNA-binding protein, to be significantly associated with the progression of ESCC. Additional immunofluorescence staining demonstrated that PURα forms cytoplasmic stress granules to suppress mRNA translation initiation. The expression level of cytoplasmic PURα in ESCC tumor tissues was significantly higher than that in adjacent epithelia and correlated with a worse patient survival rate by immunohistochemistry. Functionally, PURα strongly preferred to bind to UG-/U-rich motifs and mRNA 3´UTR by CLIP-seq analysis. Moreover, PURα knockout significantly increased the protein level of insulin-like growth factor binding protein 3 (IGFBP3). In addition, it was further demonstrated that PURα-interacting proteins are remarkably associated with translation initiation factors and ribosome-related proteins and that PURα regulates protein expression by interacting with translation initiation factors, such as PABPC1, eIF3B and eIF3F, in an RNA-independent manner, while the interaction with ribosome-related proteins is significantly dependent on RNA. Specifically, PURα was shown to interact with the mRNA 3´UTR of IGFBP3 and inhibit its expression by suppressing mRNA translation initiation. Together, this study identifies cytoplasmic PURα as a modulator of IGFBP3, which could be a promising therapeutic target for ESCC treatment.
Similar content being viewed by others
Introduction
Esophageal squamous cell carcinoma (ESCC), with a 5-year survival rate of approximately 15% to 20%, is one of the most lethal gastrointestinal malignancies worldwide [1,2,3]. Although some targeted anticancer drugs, such as gefitinib and PD-L1 blockers, have been adopted in the clinic [4], the mortality rate of ESCC is still relatively high owing to invasion and distant metastasis. Large-scale whole-exome sequencing (WES) of ESCC has identified some high-frequency gene mutations, including copy number alterations and somatic mutations [5,6,7]. In addition, epigenetic alterations in ESCC, such as DNA methylation and histone acetylation, have also been partially characterized [8]. Despite great advances in the genomic and epigenetic aspects of ESCC, the underlying mechanisms of tumor progression remain poorly understood. Hence, it is imperative to further investigate the mechanisms.
Purine-rich element binding protein alpha (PURα), encoded by PURΑ, is a single-stranded DNA/RNA-binding protein that is highly conserved from bacteria to humans [9]. PURΑ knockout in mice and PURΑ mutation in humans both result in severe neurological disease [10,11,12,13,14]. Abnormal PURα expression is also involved in the progression of several cancers, such as acute myeloid leukemia (AML) and prostate cancer [15, 16], and our previous results have shown that overexpression of PURα promotes ESCC progression [17]. Thus far, PURα has been implicated primarily in DNA replication, transcription and the cell cycle [18,19,20,21,22,23]. Recent reports have indicated that PURα in the cytoplasm encapsulates specific RNAs with some RNA-binding proteins to regulate mRNA transport [24,25,26,27,28], and emerging evidence suggests that PURα is a novel component of cytoplasmic stress granules [25, 29,30,31]. Stress granules are cytoplasmic RNA–protein complexes that form when translation initiation is limited [32, 33] and have been proposed to play an important role in neurodegenerative diseases and tumor progression [34, 35]. For example, PURα colocalized with mutant FUS in stress granules to modulate amyotrophic lateral sclerosis (ALS) pathology [25, 31]. In addition, PURα is known to associate with noncoding RNAs (e.g., TAR RNA [36], BC200 [26] and circSamd4 [37]). PURα strongly influences the development and progression of disease by regulating DNA replication, transcription and mRNA transport, but whether PURα participates in tumor progression by regulating mRNA-based processes remains unclear.
Here, we found that PURα participates in the formation of cytoplasmic stress granules and that the expression level of cytoplasmic PURα was significantly increased in ESCC tissues compared to nontumorous tissues and that ESCC patients with high expression levels of cytoplasmic PURα had a lower survival rate than those with low expression levels. We further revealed that PURα repressed the mRNA translation initiation of insulin-like growth factor binding protein 3 (IGFBP3) by forming cytoplasmic stress granules. In addition, knockdown of IGFBP3 significantly reversed the inhibitory effects of PURα loss on the cell proliferation, migration and invasion properties of KYSE170 ESCC cells. In brief, our results support that cytoplasmic PURα mediates ESCC progression by binding to the mRNA 3´UTR.
Results
Cytoplasmic PURα participates in the formation of stress granules and significantly correlates with ESCC progression
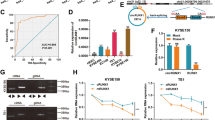
It has been commonly reported that PURα is involved in the progression of several cancers as a transcription factor [9, 18, 23]. Intriguingly, immunofluorescence staining indicated that there was the considerable cytoplasmic localization of PURα in ESCC cells and that cytoplasmic PURα was evenly dispersed as granules or accumulated around the nucleus in nongranules (Fig. 1A). Increasing evidence has reported that PURα is a core component of cytoplasmic stress granules [25, 29,30,31], suggesting that cytoplasmic PURα-positive granules in ESCC cells are likely a form of stress granules. To this end, the colocalization between PURα-positive granules and G3BP1, a well-known cytoplasmic stress granule maker [32, 33, 38], was further detected by immunofluorescence staining. It was observed that G3BP1 is localized in PURα-positive granules and that the number of PURα/G3BP1-positive granules under stress conditions markedly increased compared with those under native conditions, while the number of G3BP1-positive granules markedly decreased after the loss of PURα (Fig. 1B, C), demonstrating that cytoplasmic PURα in ESCC cells participates in the formation of stress granules. In addition, we also observed that there was more expression of PURα in the cytoplasm than in the nucleus in the ESCC tissues by immunohistochemical staining (n = 526) (Fig. 1D, E), and the protein fractionation analysis also indicated that PURα was additionally localized in the cytoplasm to a much greater extent than in the nucleus of ESCC cells (Fig. S1A). Furthermore, we compared the expression of cytoplasmic PURα in ESCC tissues (n = 282) and adjacent nontumorous epithelia (n = 282) and observed that cytoplasmic PURα expression was significantly increased in ESCC tissues (Fig. 1D, F). Importantly, Kaplan–Meier survival analysis of a total of 526 ESCC patients showed that ESCC patients with high expression levels of cytoplasmic PURα had a lower survival rate than those with low PURα (Fig. 1G), implying that cytoplasmic PURα is linked to ESCC progression. There were no correlations between PURα levels and sex, age, tumor differentiation or other factors (Supplementary Table S1).
A The localization of endogenous PURα in esophageal epithelium Het-1A and ESCC cancer (KYSE170) cells was visualized by immunofluorescence assay. PURα proteins dispersed in the cytoplasm as granules in KYSE170 cells or accumulated around the nucleus in Het1A cells. Scale bars: 30 μm. B The colocalization between endogenous PURα and the stress granule maker G3BP1 was visualized in wild-type (WT) and PURα-deficient KYSE170 (KO) cells by immunofluorescence staining. Scale bars: 30 μm. C The number of PURα/G3BP1-positive granules under stress conditions or not was calculated separately. ***p < 0.001; ns, not significant. D PURα expression in ESCC tumor tissues (first and second panels) and adjacent nontumor epithelia (third panel) was compared by immunohistochemical staining (IHC). PURα protein mainly located in cytoplasm (first panel) or nucleus (second panel) is shown. The representative region (black frame) at low magnification (40×, left) was amplified at high magnification (100×, right). Scale bars: 50 μm. E Violin plots of the statistical data regarding the IHC score of PURα protein in the cytoplasm and nucleus of tumor tissues (n = 526). ***p < 0.001 by Mann–Whitney test. F Violin plots of the statistical data regarding the IHC scores for cytoplasmic PURα in ESCC (tumor) and adjacent nontumor (normal) tissues (n = 282) were drawn. ***p < 0.001 by Mann–Whitney. G Kaplan–Meier analyses of overall survival. Patients with high cytoplasmic PURα expression (n = 296) had a significantly lower overall survival rate than patients with low cytoplasmic PURα expression (n = 230).
Extensive RNA targets of PURα in ESCC cells were revealed by CLIP-seq analysis
Cytoplasmic stress granules are ribonucleoprotein granules and are involved in the regulation of RNA homeostasis [32, 33], so we speculated that cytoplasmic PURα likely modulates ESCC progression through interaction with mRNA. Cytoplasmic PURα notably affects brain development in humans and mice as an RNA-binding protein, but the characteristics of RNA bound to PURα have not been fully elucidated [10,11,12,13,14, 24, 26,27,28]. We thus first identified RNA targets of human PURα in ESCC KYSE510 cells by CLIP-seq analysis according to reported methods [39]. In total, high-throughput sequencing yielded ~38.5 and ~34.4 million raw reads from two independent replicated PURα CLIP-seq datasets (Supplementary Table S2). After discarding low-quality raw reads and normalization, ~17.0 and ~15.2 million clean reads were generated, respectively. Of these clean reads, 82.27% (~13.9 million) and 83.52% (~12.7 million) were unambiguously mapped to the human reference genome (hg38) (Supplementary Table S3). The vast majority of uniquely mapped reads (62.98% and 57.48%) mapped to introns. Then, PURα-binding sites were predicted with a peak calling algorithm as previously reported [
Materials and methods
Crosslinking immunoprecipitation and high-throughput sequencing (CLIP-seq)
Cells were washed with ice-cold PBS 3 times and then subjected to UV crosslinking with UVC radiation (254 nm) at 400 mJ/cm2. The crosslinked cells were scraped off the plate and collected by centrifugation at 1000 × g for 5 min. The cells were lysed in cold lysis buffer (1× PBS, 0.1% SDS, 0.5% NP-40 and 0.5% sodium deoxycholate) supplemented with 200 U/ml RNase inhibitor (Takara, Kyoto, Japan) and protease inhibitor cocktail (Roche, Basel, Switzerland) for 10 min. The cell lysates were cleared by centrifugation at 10,000 rpm for 20 min at 4 °C, and the supernatants were utilized for RNase digestion and immunoprecipitation. Then, RNase T1 (Thermo Fisher Scientific, Waltham, MA, USA) was added to the lysate to a final concentration of 1 U/μl, and the mixture was incubated at 22 °C for 15 min. For immunoprecipitation, 300 μl of lysate was incubated with 10 μg of anti-PURα antibody (Abcam, Cat# ab125200) or control IgG antibody overnight at 4 °C. The immunoprecipitates were further incubated with protein A Dynabeads for 3 h at 4 °C. After collection with a magnetic field and removal of the supernatants, the beads were sequentially washed twice with wash buffer (250 mM Tris 7.4, 750 mM NaCl, 10 mM EDTA, 0.1% SDS, 0.5% NP-40 and 0.5% sodium deoxycholate) and polynucleotide kinase (PNK) buffer (50 mM Tris, 20 mM EGTA and 0.5% NP-40). Protective on-bead digestion was performed by adding MNase (Thermo Fisher Scientific) to a final concentration of 1 U/ml followed by incubation at 37 °C for 15 min. After washing with PNK buffer as described above, dephosphorylation and phosphorylation were performed with calf intestinal alkaline phosphatase (New England Biolabs, Ipswich, MA, USA) and PNK, respectively. The immunoprecipitated protein–RNA complex was eluted from the beads by heat denaturing and resolved on a Novex Bis-Tris 4–12% precast polyacrylamide gel (Thermo Fisher Scientific). The protein–RNA complexes were cut from the gel, and RNA was extracted with TRIzol after digesting the proteins. The recovered RNA was used to generate a paired-end sequencing library with a TruSeq small RNA library preparation kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. Libraries corresponding to 200–500 bp were purified, quantified and stored at −80 °C until they were used for sequencing. For high-throughput sequencing, the libraries were prepared following the manufacturer’s instructions and applied to an Illumina NextSeq 500 system for 151 bp paired-end sequencing by ABlife, Inc. (Wuhan, China).
For CLIP-seq data, adaptors and low-quality bases were trimmed from the raw sequencing reads using the FASTX-Toolkit (Version 0.0.13), and reads less than 16 nt in length were discarded. The clean reads were aligned to the human GRCH38 genome using TopHat2 with 2 mismatches [60].
After the reads were aligned onto the genome, we discarded the reads with multiple genomic locations due to ambiguous origination. Identically aligned reads were counted and merged as unique reads. The binding regions of PURα in the genome were identified using the “ABLIRC” strategy as previously reported [40]. The PURα and IgG samples were analyzed by the simulation independently. After simulation, the PURα peaks that overlapped with IgG peaks were removed. The target genes of PURα were finally determined by analyzing the locations of all the PURα binding peaks on the human genome, and the binding motifs of PURα were called with Homer software [61].
Mass spectrometry (MS) and data analysis
For protein sample preparation, the whole-cell extracts were subjected to immunoprecipitation with a specific anti-PURα antibody or rabbit IgG conjugated to magnetic beads as described in the coimmunoprecipitation assay section, and the beads destined for MS analysis were washed in 0.1% NP-40 lysis buffer with no detergent. Then, the proteins coupled to the bead-Ig complex were treated with trypsin as described previously [62].
LC–MS/MS analysis was performed using an Ultimate 3000 RSLCnano system coupled online to a Q Exactive mass spectrometer (Thermo Fisher Scientific). Four microliters of sample was injected onto a trap column (Acclaim PepMap 100, 300 μm × 5 mm, C18, 5 μm, 100 Å; flow rate 30 μl/min). Subsequently, the peptides were separated on an analytical column (Acclaim PepMap RSLC, 75 μm × 50 cm, nano Viper, C18, 2 μm, 100 Å) with a gradient of 5% to 40% solvent B over 120 min [solvent A: 0.1% formic acid (FA), solvent B: 0.1% FA, 84% acetonitrile (ACN); flow rate 400 nl/min; column oven temperature 60 °C]. The eluted peptides were analyzed online with a Q Exactive mass spectrometer using a nanoelectrospray interface. Ionization (1.8 kV ionization potential) was performed with stainless-steel emitters. The peptide ions were obtained through the following data-dependent acquisition steps: (1) a full MS scan (mass-to-charge ratio (m/z) 400 to 1800) and (2) MS/MS. The MS resolution was 70,000 at m/z 400, the automatic gain control was 3 × 106, and the maximum injection time 20 ms. For MS2, the resolution was 17,500 at m/z 400, the automatic gain control was 2 × 105, the maximum injection time 100 ms, the isolation window m/z = 2, the normalized collision energy was 27, the underfill ratio was 1%, and the intensity threshold was 2.0 × 104. The charge state was 2, and the dynamic exclusion time was 30 s.
For data analysis, the resulting MS/MS data were processed using Thermo Proteome Discoverer (PD, v2.4.1.15). The tandem mass spectra were searched against the Homo sapiens database concatenated with a reverse decoy database. Trypsin was specified as the cleavage enzyme, and up to 2 missed cleavages were allowed. The mass tolerance values for precursor ions and fragment ions were set as 10 ppm and 0.02 Da, respectively. Carbamidomethyl on Cys was specified as the fixed modification. Acetylation on the protein N-terminus and oxidation on Met were specified as the variable modifications. The peptide false discovery rate (FDR) was calculated using Percolator provided by PD. When the q value was smaller than 1%, the peptide spectrum match (PSM) was considered to be correct. Peptides assigned only to one given protein group were considered unique. The FDR was also set to 0.01 for protein identification. The peak areas of fragment ions were used to calculate the relative intensity of precursor ions for selected peptides. At least one peptide was selected for quantification of one protein. The means of the relative intensities of selected peptides represent the relative expression levels of the proteins.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8.0 software (GraphPad Software, San Diego, CA, USA). All data are presented as the mean ± SEM. Differences between two groups were compared by two-tailed Student’s t tests or the Mann–Whitney rank test. Differences among the means of three or more groups were analyzed by analysis of variance (ANOVA) followed by a multiple comparisons test. The correlation between two groups was examined using Pearson’s correlation. Survival analyses were performed by the Kaplan–Meier method and compared by the log-rank test. A p value less than 0.05 was considered to indicate statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001).