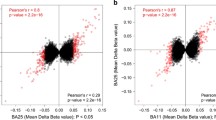
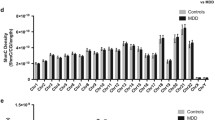
Abstract
Suicide rates have increased steadily world-wide over the past two decades, constituting a serious public health crisis that creates a significant burden to affected families and the society as a whole. Suicidal behavior involves a multi-factorial etiology, including psychological, social and biological factors. Since the molecular neural mechanisms of suicide remain vastly uncharacterized, we examined transcriptional- and methylation profiles of postmortem brain tissue from subjects who died from suicide as well as their neurotypical healthy controls. We analyzed temporal pole tissue from 61 subjects, largely free from antidepressant and antipsychotic medication, using RNA-sequencing and DNA-methylation profiling using an array that targets over 850,000 CpG sites. Expression of NPAS4, a key regulator of inflammation and neuroprotection, was significantly downregulated in the suicide decedent group. Moreover, we identified a total of 40 differentially methylated regions in the suicide decedent group, map** to seven genes with inflammatory function. There was a significant association between NPAS4 DNA methylation and NPAS4 expression in the control group that was absent in the suicide decedent group, confirming its dysregulation. NPAS4 expression was significantly associated with the expression of multiple inflammatory factors in the brain tissue. Overall, gene sets and pathways closely linked to inflammation were significantly upregulated, while specific pathways linked to neuronal development were suppressed in the suicide decedent group. Excitotoxicity as well as suppressed oligodendrocyte function were also implicated in the suicide decedents. In summary, we have identified central nervous system inflammatory mechanisms that may be active during suicidal behavior, along with oligodendrocyte dysfunction and altered glutamate neurotransmission. In these processes, NPAS4 might be a master regulator, warranting further studies to validate its role as a potential biomarker or therapeutic target in suicidality.
Similar content being viewed by others
Introduction
Suicide is a leading cause of death, with more than 700,000 cases registered across the globe each year [1]. Suicide tragically affects all age groups, including adolescents, pregnant and post-partum women, as well as elderly individuals. It is the most common cause of death due to non-accidents for people below the age of 35 years [2]. Mood disorders such as major depressive disorder (MDD) are commonly associated with suicide [3], with as many as 60% of those who die by suicide having had a diagnosis of MDD [4]. MDD has an elevated rate of both suicidal ideation, and nonfatal and fatal suicide attempts [66]. Interestingly, HSPA1B and HSPA1A were ranked 60 and 53 by the lowest P-value in our DE gene list, while GNAS was a differentially methylated gene in our suicide decedent group. Moreover, estrogen signaling was also close to significant in our transcriptome enrichment analysis (KEGG pathway analysis, FDR = 0.053).
As our next step, we performed an enrichment analysis, which showed that several gene sets and pathways related to inflammation, infection, and neuronal development were both differentially expressed and methylated in the suicide decedents. Overall, the suicide decedent group showed an activation of gene sets related to inflammation and excitotoxic mechanisms, accompanied by a suppression of gene sets related to maturation of OLs and myelination. The deconvolution analysis indicated an increased ratio of inhibitory neurons against excitatory neurons in the suicide decedent group and further an imbalance in several neurotransmitters, as illustrated in Supplementary Fig. 1. The balance of inhibitory and excitatory neurotransmitter levels plays a significant role in several psychiatric disorders including MDD [67, 68]. Interestingly, the concentration of these neurotransmitters might not only be associated with the numbers of their interneurons but also with their functions [69, 70].
Moreover, utilizing deconvolution analysis, we found that the suicide decedent group had fewer OLs than the healthy control group in all reference brain regions. In line with this, Aston and colleagues have previously reported abnormalities of oligodendroglia in the temporal pole from patients with MDD by transcriptome analysis [71]. Our enrichment analysis demonstrated that the suicide decedent group exhibited suppression of a gene set that was also found to be down regulated in Aston’s study in MDD (Fig. 2A). Our enrichment analysis identified 10 gene sets and pathways that relate to OLs development and myelination process. Genes encoding the key myelination-related proteins Myelin Basic Protein (MBP) and Myelin Associated Oligodendrocyte Basic protein (MOBP) ranked 11th and 25th in our DE gene list. Consistent with our findings, previous transcriptome analyses of suicide postmortem frontal cortex tissue found a lower expression of genes involved in OLs differentiation [19] and a reduction of OPCs in subjects who died from a violent suicide compared to a non-violent suicide [64]. We also found the gene ZNF24 to be significantly hypermethylated in the suicide decedent group. ZNF24 (also known as ZFP24, ZFP191) is a transcriptional regulator, and phosphorylation of ZNF24 controls the developmental process of oligodendrocyte progenitor cells to pre-myelinating OLs [72] and as such it is required by OLs during myelination [73]. We detected that almost all the ZNF24 CpGs were located in the promotor region and hypermethylated in the suicide decedent group, consistent with suppression of gene expression [74].
Previous studies have shown that OLs and their precursors are vulnerable to inflammation, oxidative stress and elevated glutamate levels [75], which explains why these cells are affected in multiple neuropathological entities, including Alzheimer’s disease, spinal cord injury, Parkinson’s disease, and ischemia, as well as during hypoxia [76, 77]. OL deficits have also been previously reported in schizophrenia and bipolar disorder [78, 79] and direct evidence of altered oxidative stress markers has been detected in OLs from MDD suicide postmortem brain tissues [80]. The coincidence of gene expression patterns between upregulation of immune response and down regulation of maturation of OLs in suicide decedents in our study might suggest a causal relationship between the two. Indeed, OPCs were shown to be cytotoxic targets of neuroinflammation in a study of the demyelinating disease multiple sclerosis [81]. Also, there is experimental evidence that OLs block their own differentiation in response to inflammation by activating toll-like receptor-3 (TLR3) [82]. Deconvolution analysis heavily relies on the reference datasets used. While we utilized a widely cited single-cell sequencing dataset from the human brain [44], it is worth noting that the brain regions in this dataset do not precisely align with Brodmann Area 20, as we were unable to find a single-cell dataset specific only to this region. However, in the regions utilized for deconvolution, there is a partial overlap with Brodmann Area 20, and we were able to identify a consistent pattern of lower oligodendrocyte cell populations in the suicide decedent group compared to the control group. To further validate our findings and address the regional specificity, conducting future single-cell RNA sequencing specifically from Brodmann Area 20 would be valuable.
Another recent integrative DNA methylation (via Infinium human 450 BeadChip) and gene expression analysis (via Illumina HumanHT-12 V4 Expression BeadChip) on postmortem brain tissue from male suicide decedents identified 622 differentially expressed genes in the suicide decedent group compared with controls [83]. Among them, 70 genes had concordant methylation and expression changes including genes relevant to psychiatric disorders such as ADCY9, CRH, NFATC4. None of these genes were differentially expressed or methylated in our study. That study differs from our current one in that more than half of the subjects in the previous study had a history of substance abuse, and many, including the controls, had a variety of psychiatric disorders [83]. In contrast, the suicide decedents in our study were largely free from psychotrophic medication, as confirmed with postmortem toxicology, and had a confirmed diagnosis of MDD. The region of analysis, the prefrontal cortex, was also different from our current study as we utilized tissue from the temporal pole, another region proposed to be involved in suicidal behavior [84, 85]. We have previously conducted a pilot study analyzing prefrontal cortical tissue and found evidence of both hypermethylation and a focus of findings in inflammation and neurotrophic pathways [86, 87].
Limitations of this study
Our DNA methylation analysis used Illumina EPIC array platform which is designed to cover 30% of the human methylome [88]. Thus, inferences made from these methylation profiles require cautious interpretation. Moreover, our DNA methylation analysis only detects the CpG sites while it has been shown that non-CpG methylation can also play a role in both neurons and glial cells [89], especially later in life [90]. Furthermore, our sample sizes are relatively small for the Illumina Epic 850k array, so it is likely that we have missed additional differentially methylated regions [91]. Another limitation is the dissection process of the brain tissue, which can lead to slight variation in the anatomical region used for analysis. Since the dissected brain gyri fold in three dimensions, it is impossible to exclude all white matter while still taking the full thickness (~ 3 mm) of the cortical ribbon. However, it is important to note that any random variation during dissection should not impact the groups differently, as pathologists were blind to future study design and analytical approaches. Despite the limitations of this study, it is important to note that the data presented here offer important indications of distinct molecular signatures in well-characterized suicidal individuals, without any major influence of psychotrophic medication, and that they can serve as the basis for designing future targeted studies.
Conclusions
Overall, our study suggests a network of mechanisms involved in suicidal behavior, centering on upregulated inflammatory pathways and the suppression of oligodendrocyte-related genes. It is plausible to propose that in the brain, NPAS4 may serve as a master transcriptional regulator that modulates neural and neuronal circuit development while maintaining mitochondrial and immune function. NPAS4 downregulation could lead to excitatory and inhibitory imbalance, impaired neural development, increased oxidative stress and neuroinflammation. Our results confirm the involvement of inflammatory pathways in active suicidal behavior and suggest that NPAS4-associated mechanisms might serve as novel targets in the development of therapies for suicide prevention. Further, this work also supports that the role of OLs should be further evaluated in suicidal behavior.
Code availability
The codes used in this project can be found at https://github.com/psychesha21/RNAseq_Analysis and https://github.com/psychesha21/DNA-methylation. Sequencing data for RNA and DNA methylation can be accessed by GEO accession number: SuperSeries GSE243488.
References
World Health Organization. Facts sheet: suicide. https://www.who.int/news-room/fact-sheets/detail/suicide.
National Institute of Mental Health. Suicide. https://www.nimh.nih.gov/health/statistics/suicide.
Nock MK, Hwang I, Sampson N, Kessler RC, Angermeyer M, Beautrais A, et al. Cross-national analysis of the associations among mental disorders and suicidal behavior: findings from the WHO world mental health surveys. PLoS Med. 2009;6:e1000123.
Tanney BL Psychiatric diagnoses and suicidal acts. In: RW Maris, AL Berman, MM Silverman, editors. Comprehensive textbook of suicidology, New York: Guilford Press; 2000. 311–41.
Cai H, **e X-M, Zhang Q, Cui X, Lin J-X, Sim K, et al. Prevalence of suicidality in major depressive disorder: a systematic review and meta-analysis of comparative studies. Front Psych. 2021;12:690130.
Turecki G, Brent DA, Gunnell D, O’Connor RC, Oquendo MA, Pirkis J, et al. Suicide and suicide risk. Nat Rev Dis Prim. 2019;5:74.
Soreff SM, Basit H, Attia FN Suicide risk. Treasure Island: StatPearls Publishing; 2021.
Diekstra RFW, Garnefski N. On the nature, magnitude, and causality of suicidal behaviors: an international perspective. Suicide Life Threat Behav. 1995;25:36–57.
Voracek M, Loibl L. Genetics of suicide: a systematic review of twin studies. Wien Klin Wochenschr. 2007;119:463–75.
Ruderfer DM, Walsh CG, Aguirre MW, Tanigawa Y, Ribeiro JD, Franklin JC, et al. Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Mol Psych. 2020;25:2422–30.
Mullins N, Bigdeli TB, Børglum AD, Coleman JRI, Demontis D, Mehta D, et al. GWAS of suicide attempt in psychiatric disorders and association with major depression polygenic risk scores. Am J Psych. 2019;176:651–60.
Willour VL, Seifuddin F, Mahon PB, Jancic D, Pirooznia M, Steele J, et al. A genome-wide association study of attempted suicide. Mol Psych. 2012;17:433–44.
Galfalvy H, Zalsman G, Huang Y-Y, Murphy L, Rosoklija G, Dwork AJ, et al. A pilot genome wide association and gene expression array study of suicide with and without major depression. World J Biol Psych. 2013;14:574–82.
Black C, Miller BJ. Meta-analysis of cytokines and chemokines in suicidality: distinguishing suicidal versus nonsuicidal patients. Biol Psych. 2015;78:28–37.
Lindqvist D, Janelidze S, Hagell P, Erhardt S, Samuelsson M, Minthon L, et al. Interleukin-6 is elevated in the cerebrospinal fluid of suicide attempters and related to symptom severity. Biol Psych. 2009;66:287–92.
Enache D, Pariante CM, Mondelli V. Markers of central inflammation in major depressive disorder: a systematic review and meta-analysis of studies examining cerebrospinal fluid, positron emission tomography and post-mortem brain tissue. Brain Behav Immun. 2019;81:24–40.
Van Heeringen K, Mann JJ. The neurobiology of suicide. Lancet Psych. 2014;1:63–72.
Pandey G, Rizavi H, Bhaumik R, Ren X. Innate immunity in the postmortem brain of depressed and suicide subjects: role of Toll-like receptors. Brain Behav Immun. 2018;75:101–11.
Pantazatos SP, Huang Y-Y, Rosoklija GB, Dwork AJ, Arango V, Mann JJ. Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Mol Psych. 2017;22:760–73.
Policicchio S, Washer S, Viana J, Iatrou A, Burrage J, Hannon E, et al. Genome-wide DNA methylation meta-analysis in the brains of suicide completers. Transl Psych. 2020;10:69.
Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, et al. Differential glucocorticoid receptor exon 1B, 1C, and 1H expression and methylation in suicide completers with a history of childhood abuse. Biol Psych. 2012;72:41–8.
Guintivano J, Brown T, Newcomer A, Jones M, Cox O, Maher BS, et al. Identification and replication of a combined epigenetic and genetic biomarker predicting suicide and suicidal behaviors. Am J Psych. 2014;171:1287–96.
Underwood MD, Bakalian MJ, Escobar T, Kassir S, Mann JJ, Arango V. Early-life adversity, but not suicide, is associated with less prefrontal cortex gray matter in adulthood. Int J Neuropsychopharmacol. 2019;22:349–57.
Spitzer RL, Williams JBW, Gibbon M, First MB. The structured clinical interview for DSM-III-R (SCID): I: history, rationale, and description. Arch Gen Psych. 1992;49:624–9.
First M, Gibbon M, Spitzer R, Benjamin L, Williams J. Structured clinical interview for DSM-IV axis II personality disorders (SCID-II). Washington DC: American Psychiatric Press, 1997.
Brown GL, Goodwin FK, Ballenger JC, Goyer PF, Major LF. Aggression in humans correlates with cerebrospinal fluid amine metabolites. Psych Res. 1979;1:131–9.
Oquendo MA, Halberstam B, Mann JJ. Risk factors for suicidal behavior: Utility and limitations of research instruments. In: First MB. editor. Standardized Evaluation in Clinical Practice. American Psychiatric Publishing, Inc.; 2003. pp. 103–130.
Kelly TM, Mann JJ. Validity of DSM-III-R diagnosis by psychological autopsy: a comparison with clinician ante-mortem diagnosis. Acta Psych Scand. 1996;94:337–43.
Clive ML, Boks MP, Vinkers CH, Osborne LM, Payne JL, Ressler KJ, et al. Discovery and replication of a peripheral tissue DNA methylation biosignature to augment a suicide prediction model. Clin Epigenetics. 2016;8:113.
Martin M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Wang L, Nie J, Sicotte H, Li Y, Eckel-Passow JE, Dasari S, et al. Measure transcript integrity using RNA-seq data. BMC Bioinforma. 2016;17:58.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–50.
Igor Dolgalev. msigdbr: MSigDB gene sets for multiple organisms in a tidy data format. 2022.
Venables WN, Ripley BD. Modern applied statistics with S. 4th ed. Springer; 2002.
McKenzie AT, Katsyv I, Song W-M, Wang M, Zhang B. DGCA: a comprehensive R package for differential gene correlation analysis. BMC Syst Biol. 2016;10:106.
Miller HE, Bishop AJR. Correlation AnalyzeR: functional predictions from gene co-expression correlations. BMC Bioinforma. 2021;22:206.
New York Genome Center. https://www.nygenome.org/.
Zhou W, Triche TJ Jr, Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucl Acids Res. 2018;46:e123.
Ren X, Kuan PF. methylGSA: a Bioconductor package and Shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics. 2019;35:1958–9.
Dong M, Thennavan A, Urrutia E, Li Y, Perou CM, Zou F, et al. SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Brief Bioinform. 2021;22:416–27.
Wang X, Park J, Susztak K, Zhang NR, Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10:380.
Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–8.
The Allen Institute. Cell type database: RNA-Seq data. https://portal.brain-map.org/atlases-and-data/rnaseq.
Brooks M, Kristensen K, van Benthem K, Magnusson A, Berg CW, Nielsen A, et al. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 2017;9:378–400.
Russell V Lenth. emmeans: estimated marginal means, aka least-squares means. 2022. https://CRAN.R-project.org/package=emmeans.
Koller M. robustlmm: an R package for robust estimation of linear mixed-effects models. J Stat Softw. 2016;75:1–24.
Coutellier L, Beraki S, Ardestani PM, Saw NL, Shamloo M. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS One. 2012;7:e46604.
Fu J, Guo O, Zhen Z, Zhen J. Essential functions of the transcription factor Npas4 in neural circuit development, plasticity, and diseases. Front Neurosci. 2020;14:603373.
Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–5.
Choy FC, Klarić TS, Koblar SA, Lewis MD. The role of the neuroprotective factor Npas4 in cerebral ischemia. Int J Mol Sci. 2015;16:29011–28.
Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–29.
Choy FC, Klarić TS, Leong WK, Koblar SA, Lewis MD. Reduction of the neuroprotective transcription factor Npas4 results in increased neuronal necrosis, inflammation and brain lesion size following ischaemia. J Cereb Blood Flow Metab. 2015;36:1449–63.
Hughes BW, Siemsen BM, Tsvetkov E, Berto S, Kumar J, Cornbrooks RG, et al. NPAS4 in the medial prefrontal cortex mediates chronic social defeat stress-induced anhedonia-like behavior and reductions in excitatory synapses. Elife. 2023;12:e75631.
Jaehne EJ, Klarić TS, Koblar SA, Baune BT, Lewis MD. Effects of Npas4 deficiency on anxiety, depression-like, cognition and sociability behaviour. Behavioural Brain Res. 2015;281:276–82.
Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–75.
Sosnowski DW, Jaffe AE, Tao R, Deep-Soboslay A, Shu C, Sabunciyan S, et al. Differential expression of NPAS4 in the dorsolateral prefrontal cortex following opioid overdose. Drug Alcohol Depend Rep. 2022;3:100040.
Zhang S-Y, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, et al. Inborn errors of RNA lariat metabolism in humans with brainstem Viral Infection. Cell. 2018;172:952–65.e18.
Antico Arciuch VG, Tedesco L, Fuertes M, Arzt E. Role of RSUME in inflammation and cancer. FEBS Lett. 2015;589:3330–5.
Mellström B, Sahún I, Ruiz-Nuño A, Murtra P, Gomez-Villafuertes R, Savignac M, et al. DREAM controls the on/off switch of specific activity-dependent transcription pathways. Mol Cell Biol. 2014;34:877–87.
Benito E, Barco A. The neuronal activity-driven transcriptome. Mol Neurobiol. 2015;51:1071–88.
Usami S, Nishio S. Nonsyndromic hearing loss and deafness, mitochondrial. University of Washington, Seattle, Seattle (WA); 2004.
Punzi G, Ursini G, Chen Q, Radulescu E, Tao R, Huuki LA, et al. Genetics and brain transcriptomics of completed suicide. Am J Psych. 2022;179:226–41.
Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Zhao Y, Wang L, Wu Y, Lu Z, Zhang S. Genome-wide study of key genes and scoring system as potential noninvasive biomarkers for detection of suicide behavior in major depression disorder. Bioengineered. 2020;11:1189–96.
Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry. 2011;16:383–406.
Godfrey KEM, Gardner AC, Kwon S, Chea W, Muthukumaraswamy SD. Differences in excitatory and inhibitory neurotransmitter levels between depressed patients and healthy controls: a systematic review and meta-analysis. J Psychiatr Res. 2018;105:33–44.
Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315.
Filice F, Vörckel KJ, Sungur AÖ, Wöhr M, Schwaller B. Reduction in parvalbumin expression not loss of the parvalbumin-expressing GABA interneuron subpopulation in genetic parvalbumin and shank mouse models of autism. Mol Brain. 2016;9:10.
Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psych. 2005;10:309–22.
Elbaz B, Aaker JD, Isaac S, Kolarzyk A, Brugarolas P, Eden A, et al. Phosphorylation state of ZFP24 controls oligodendrocyte differentiation. Cell Rep. 2018;23:2254–63.
Howng SYB, Avila RL, Emery B, Traka M, Lin W, Watkins T, et al. ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev. 2010;24:301–11.
Anastasiadi D, Esteve-Codina A, Piferrer F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin. 2018;11:37.
Haroon E, Miller AH, Sanacora G. Inflammation, glutamate, and glia: a trio of trouble in mood disorders. Neuropsychopharmacology. 2017;42:193–215.
Spaas J, van Veggel L, Schepers M, Tiane A, van Horssen J, Wilson DM, et al. Oxidative stress and impaired oligodendrocyte precursor cell differentiation in neurological disorders. Cell Mol Life Sci. 2021;78:4615–37.
Nasrabady SE, Rizvi B, Goldman JE, Brickman AM. White matter changes in Alzheimer’s disease: a focus on myelin and oligodendrocytes. Acta Neuropathol Commun. 2018;6:22.
Valdés-Tovar M, Rodríguez-Ramírez AM, Rodríguez-Cárdenas L, Sotelo-Ramírez CE, Camarena B, Sanabrais-Jiménez MA, et al. Insights into myelin dysfunction in schizophrenia and bipolar disorder. World J Psych. 2022;12:264–85.
Tkachev D, Mimmack M, Ryan M, Wayland M, Freeman T, Jones P, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805.
Chandley MJ, Szebeni A, Szebeni K, Wang-Heaton H, Garst J, Stockmeier CA, et al. Markers of elevated oxidative stress in oligodendrocytes captured from the brainstem and occipital cortex in major depressive disorder and suicide. Prog Neuropsychopharmacol Biol Psych. 2022;117:110559.
Kirby L, ** J, Cardona JG, Smith MD, Martin KA, Wang J, et al. Oligodendrocyte precursor cells present antigen and are cytotoxic targets in inflammatory demyelination. Nat Commun. 2019;10:3887.
Boccazzi M, Van Steenwinckel J, Schang A-L, Faivre V, Le Charpentier T, Bokobza C, et al. The immune-inflammatory response of oligodendrocytes in a murine model of preterm white matter injury: the role of TLR3 activation. Cell Death Dis. 2021;12:166.
Romero-Pimentel AL, Almeida D, Muñoz-Montero S, Rangel C, Mendoza-Morales R, Gonzalez-Saenz EE, et al. Integrative DNA methylation and gene expression analysis in the prefrontal cortex of Mexicans who died by suicide. Int J Neuropsychopharmacol. 2021;24:935–47.
Chen C, Chen W, Zhang B. Functional alterations of the suicidal brain: a coordinate-based meta-analysis of functional imaging studies. Brain Imaging Behav. 2022;16:291–304.
Sarkinaite M, Gleizniene R, Adomaitiene V, Dambrauskiene K, Raskauskiene N, Steibliene V. Volumetric MRI analysis of brain structures in patients with history of first and repeated suicide attempts: a cross sectional study. Diagnostics (Basel). 2021;11:488.
Haghighi F, Wang Z, Chatterton Z, Schnieder T, Wilson C, Rosoklija G, et al. Biosignatures of stress in suicide neuropathology. Biol Psych. 2020;87:S145–S146.
Haghighi F, Liu Q, Sun S, Wang Z, Ge Y, Huang Y, et al. Systemic inflammation positively correlates with high suicide ideation in blood and CNS. Biol Psych. 2022;91:S370–S371.
Shu C, Zhang X, Aouizerat BE, Xu K. Comparison of methylation capture sequencing and Infinium MethylationEPIC array in peripheral blood mononuclear cells. Epigenetics Chromatin. 2020;13:51.
Jang HS, Shin WJ, Lee JE, Do JT. CpG and non-CpG methylation in epigenetic gene regulation and brain function. Genes (Basel). 2017;8:148.
Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science (1979). 2013;341:1237905.
University of Essex. EPIC array power calculations. https://epigenetics.essex.ac.uk/shiny/EPICDNAmPowerCalcs/.
Acknowledgements
The study was funded by the National Institutes of Mental Health (NIMH R01 MH118211-01A1) awarded to the multi-PI team of LB, JJM and EDA. We wish to thank all the families of the deceased participants for their donation of brain tissue, as well as the sharing of detailed clinical information by participating in our interviews. Moreover, we wish to thank the genomics core at Van Andel Institute, headed by Ms. Marie Adams, for their work on the RNA-sequencing analysis.
Author information
Authors and Affiliations
Contributions
LB, JJM and EDA jointly planned the experiments and led the analysis and interpretation of the data, as well as manuscript writing. QS performed the statistical and bioinformatical analysis of the data and drafted the first version of the manuscript. ZF contributed to the bioinformatical analysis and manuscript writing. MLEG provided advice on the experimental plan and assisted in manuscript writing, ZM provided statistical advice. JAS assisted with manuscript writing, IRB related paperwork and project logistics. HG provided statistical advice and assisted in manuscript writing. AD, GR, MDU and NS all contributed with gathering clinical information from psychological autopsies as well as neuropathological examinations and assisted with manuscript writing. All authors have contributed to, and approved of the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
EDA receives grants or contracts from Alkermes, Astellas, Biogen, Boehringer-Ingelheim, InnateVR, Janssen, Karuna, Lundbeck, National Network of Depression Centers, Neurocrine Biosciences, Otsuka, Pear Therapeutics, Takeda, Teva. He receives consulting fees from: Alkermes, Atheneum, CAPNOS Zero (unpaid), Indivior, Karuna, Lundbeck, Otsuka, Neurocrine Biosciences, Sunovion, Teva. He receives payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Otsuka/Lundbeck. He receives support for attending meetings and/or travel from Alkermes, Karuna, Otsuka/Lundbeck, Neurocrine Biosciences. Stock or stock options: AstraZeneca, Johnson and Johnson, Moderna, Pfizer. AJD receives funding from NIMH (R0112530) and the Bay Area Lyme Biobank. HG and her family own stocks in Ilumina, Inc. JJM receives royalties for commercial use of the C-SSRS from the Research Foundation for Mental Hygiene. Other co-authors have no competing interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sha, Q., Fu, Z., Escobar Galvis, M.L. et al. Integrative transcriptome- and DNA methylation analysis of brain tissue from the temporal pole in suicide decedents and their controls. Mol Psychiatry 29, 134–145 (2024). https://doi.org/10.1038/s41380-023-02311-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-023-02311-9
- Springer Nature Limited