Abstract
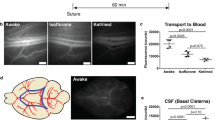
Meningeal lymphatic vessels (mLVs) play a pivotal role in regulating metabolic waste from cerebrospinal fluid (CSF). However, the current limitations in field of view and resolution of existing imaging techniques impede understanding the stereoscopic morphology and dynamic behavior of mLVs in vivo. Here, we utilized dual-contrast functional photoacoustic microscopy to achieve wide-field intravital imaging of the lymphatic system, including mLVs and glymphatic pathways. The stereoscopic photoacoustic microscopy based on opto-acoustic confocal features has a depth imaging capability of 3.75 mm, facilitating differentiation between mLVs on the meninges and glymphatic pathways within the brain parenchyma. Subsequently, using this imaging technique, we were able to visualize the dynamic drainage of mLVs and identify a peak drainage period occurring around 20–40 min after injection, along with determining the flow direction from CSF to lymph nodes. Inspiringly, in the Alzheimer’s disease (AD) mouse model, we observed that AD mice exhibit a ~ 70% reduction in drainage volume of mLVs compared to wild-type mice. With the development of AD, there is be continued decline in mLVs drainage volume. This finding clearly demonstrates that the AD mouse model has impaired CSF drainage. Our study opens up a horizon for understanding the brain’s drainage mechanism and dissecting mLVs-associated neurological disorders.
Similar content being viewed by others
Introduction
The central nervous system (CNS) has long been considered an ‘immune privilege’ organ lacking lymphatic vessels that transport immune cells1,2,3. However, recent studies on intracranial clearance have revealed the presence of meningeal lymphatic vessels (mLVs) and the glymphatic pathway for interstitial fluids and cerebrospinal fluid (CSF) drainage4,5,6,7,8. The mLVs continuously drain macromolecules, wastes, and superfluous liquids from the CNS to the periphery by connecting deep cervical lymph nodes (cLNs) and play an important role in the active transport of immune cells9,10,11. Among them, permeable collecting lymphatics and capillary lymphatics are distributed along the sigmoid sinus and petrosquamous sinus of the skull base. The lymphatic vessels located in the meninges are distributed along the transverse sinus (TS) and superior sagittal sinus (SSS)12,13,14,15. It is important to identify and characterize the routes responsible for CNS lymphatic drainage because paths of these may offer a valuable approach to the treatment of CNS-associated neuroinflammatory conditions.
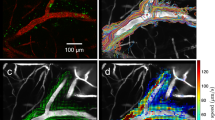
Disruption of the mLVs and cerebral vessels (CVs) have been reported to occur and contribute to the disease progression during aging as well as in neurodegenerative and neurological diseases, such as Alzheimer’s disease (AD)16,17,18,19, Parkinson’s disease20, traumatic brain injury21,22,23, encephalitis24,25, and brain tumor26,27. Differences in the morphology and function of mLVs and CVs based on sex and age may also be related to sex and age differences in the incidence of certain neurological diseases28,29,49,50 and hemodynamic and morphological variations in both superficial and deep tissues51,52,53,54. Furthermore, various photoacoustic system has been explored in brain metabolism32,55, brain injury56,57,58,59, state connectivity60, and brain response61,62 by employing different laser illumination and detection schemes. These technologies have been achieved in the study of in vivo high-resolution imaging of the brain, whereas they almost concentrate on CVs rather than mLVs. Of course, the imaging of mLVs should also be studied. Partial studies have been made in some pure-optical ways, IF way, as well as MRI way, but it is hard to visualize the structure and function of mLVs with high resolution in vivo. DCF-PAM allows simultaneous visualization of CVs and mLVs with high resolution in vivo. Vascular organizations are spread throughout the whole view due to the significant signal of hemoglobin, and mLVs are typically characterized by concentrated near the TS and SSS. The images of CVs and mLVs feature different experimental characteristics, reflecting the differences in their organizations. We hope that with the system provided in this paper, accurate characterization of CVs and mLVs in vivo will be feasible. DCF-PAM enables stereoscopic images of CVs and mLVs in vivo. Through the self-developed hollow ultrasound transducer63,64 and light-acoustic confocal system design, the obtained stereoscopic images can maintain a good signal-to-noise ratio and resolution within a depth range of 3.75 mm.
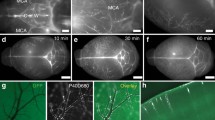
Using mLVs images analyzed, we notice that the probes are especially important in the imaging process of mLVs. The OVA-ICG as a tracer used is a macromolecule in its form. This means that probes come from CSF by drainage and clearance, and not enough covered entire mLVs to perform a detailed visualization of mLVs. All illuminated signal points tend to only show the trajectories of the probe in all mLVs. As such, labeled probes should be used to label the entire network of mLVs in the future. One can note that, even though the mouse needs to euthanize in the IF imaging, the conjugates of antibody and fluorescent dye are performed as labeled probes, and that should be taken into account when synthesizing PA probes. If labeled PA probes will be synthesized and injected into CM, the final reconstructed images may reveal entire mLVs.
For the DCF-PAM, the device based on the two-dimension motorized translation stage has a wide-view scanning range. This makes the image obtained to cover the whole brain of a mouse. In vivo, wide-view monitoring seems to include more structured information about the sample, whereas CVs and mLVs have many real-time features of flow behavior. The duration of one PA imaging in the brain of DCF-PAM does not allow the real-time representation of CVs and mLVs as it needs long acquisition times. The scanning speed is critical to the real-time recording of a series of dynamic changes. If the system in the strategy is able to use some high-speed scanning device, such as a 2-axis fast galvanometer scanner, providing high-frame-rate scanned images and realizing real-time whole-brain recording results will be possible. Our previous study has a dual-raster-scanned PA microscope that integrates a two-dimensional motorized translation stage for large field-of-view imaging and a 2-axis fast galvanometer scanner for real-time imaging65. In the future, we will have chosen this system to update the dual-wavelength system. The idea appears much better than those of many other methods. It will improve the applicability of the strategy by allowing wide-view monitoring and real-time recording.
In summary, we have proposed an approach for high-resolution image CVs and mLVs in vivo using DCF-PAM. When the advent of relative brain diseases or injury, the system devised may also help to compare and characterize such diagnoses and treatment of diseases. The use of the system is seen potential to increase new imaging technology and method and realize better and faster in vivo imaging of CVs and mLVs. It will facilitate brain study that relevant aspects of the nervous system and immune system by researchers, and allow them to take advantage of the existing condition and develop new application directions.
Materials and methods
Animal subjects
Young (1–2 months old) and adult (5–6 months, 10–12 months old) male C57BL/6 J WT mice are purchased from Guangdong Medical Laboratory Animal Center. The transgenic mice (5xFAD) are provided by Sun Yat-sen Memorial Hospital, Sun Yat-sen University. All experimental mice are of a C57BL/6 background, and WT and AD mice are paired from the litters and housed under the same living conditions. In-housed bred male transgene carriers and non-carrier (WT) littermates are used at different ages. Mice are housed in a temperature and humidity-controlled room, maintained in a 12 h/12 h light/dark cycle. All mice are fed with regular rodent chow and sterilized tap water ad libitum. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of Hainan University (approval number: HNUAUCC-2021-00112).
Cisterna magna Injection
Mice are anaesthetized by inhaling 2% isoflurane air. The hair of the neck is shaved and cleaned with iodine and 70% ethanol, an ophthalmic solution is placed on the eyes to prevent drying, and the head of the mouse is secured in a stereotaxic frame. After making a skin incision, the muscle layers are retracted, and the CM is exposed. Using a Hamilton syringe (coupled to a 30-gauge needle), the volume of desired tracer solution is injected into the CSF-filled cisterna magna compartment at a rate of about 2.5 μl per minute66. After injecting, the syringe is left in place for at least 2 min to prevent backflow of CSF. The neck skin is then sutured. The method of intra-cisterna magna injection is used to administer 10 μl of OVA-ICG. The tracers are imaged at the indicated time points and analyzed.
Photoacoustic Imaging
For PA imaging of the mouse brain, the hair of the normal mouse is first removed using a shaver and depilatory cream. Then, the mouse is removed scalp. The skull is kept intact. Imaging is performed with the scanning range of 12 × 12 × 3.75 mm3, and fast-axis scanning speed of 10 mm s–1, and the scanning time is about 12 min. During scanning, to avoid motion artifacts in living mice, firstly, the degree of anesthesia of the mice is controlled through gas anesthesia, and analgesia and central muscle relaxation medication are administered to reduce the impact of breathing. Secondly, during the process of acquiring images, the bandpass filter is used to smooth the PA signal, and median filtering is performed on every 4 A-line data to eliminate the noise. Finally, after acquiring the 3D PA data, the motion correction algorithm and bilinear interpolation are applied to further reduce motion artifacts in the image67.
In the phantom experiments, the 30-gauge needle is inserted into the mouse brain in vitro. To measure the penetration depths, 2500 B-scan images are acquired, and each B-scan image contained 500 A-lines. To evaluate the lateral resolution, the edge spread function as well as the corresponding line spread function are calculated perpendicular to the blade edge. To evaluate the axial resolution, the acquired A-line signals are performed Hilbert transformation and measured the full-width-at-half-maximum. 2500 B-scans to cover the FOV are acquired as a set of coronal views, and MAP to each B-scan image is carried out so that there were 2500× 1000 pixels in total in one image of the horizontal view. In sequential imaging experiments, PA images are continuously acquired for approximately 2 hours post-injection.
MRI acquisitions
All MRI acquisitions are performed in a 9.4 T BioSPEC system (Bruker, Germany). The mouse is placed in the MRI apparatus and maintained under light anesthesia with isoflurane (1–1.25% in oxygen). Before and after injecting DTPA-Gd into the CSF, a series of post-contrast T1-weighted images are taken through the head with the following parameters: TR = 500 ms, TE = 6 ms, FOV = 20× 20 mm2, slice thickness = 0.7 mm, number of slices = 17 and NEX = 2. The total acquisition time is about 147 s.
Immunohistochemistry and Imaging
Mice are euthanized by intraperitoneal (i.p.) injection with an overdose of anesthesia (20 mg kg–1 pentobarbital sodium) and transcardially perfused with PBS. The skin is removed from the head and the muscle is stripped from the bone. Then, the mandibles and skull rostral to maxillae are removed. Whole-mount skullcap is fixed while still attached to the skull cap in 4% paraformaldehyde for 12 h at room temperature. The meninges are then dissected from the skullcap. The tissue is rinsed in PBS and 0.5% Triton X-100 containing 5% bovine serum albumin for 1 h at room temperature. This blocking step is followed by incubation with appropriate dilutions of primary antibodies: anti-LYVE-1-Alexa Fluor 488 (eBioscience, 1:200), anti-CD31 (Millipore, MAB1398Z, 1:200) in PBS, and 0.5% Triton X-100 overnight at 4 °C. Meningeal whole-mounts are then washed three times for 10 min at room temperature in PBS and 0.5% Triton X-100 followed by incubation with appropriate Alexa Fluor 555 (Thermo Fisher Scientific, 1:400) in PBS and 0.5% Triton X-100 overnight at 4°C. Alternatively, incubate overnight at 4°C with the appropriate Anti-β-amyloid, 1-16 Antibody (Biolegend, 1:400), Alexa Fluor 647 (Thermo Fisher Scientific, 1:400). After incubating, the tissue is washed three times for 10 min with PBS and 0.5% Triton X-100 at room temperature and mounted with an IF sealing agent and glass coverslips68. Preparations are stored at 4°C for no more than one week until images are acquired using a confocal microscope (LSM 880 Confocal Laser Scanning Microscope, Zeiss).
Laser speckle
Mice are anesthetized by isoflurane. An incision is done along the midline to separate the skin of the skull and laser speckle is used to detect mice’s cerebral blood flow. Laser speckle blood flow images are recorded and used to identify the regions of interest. Within these regions, the mean blood flow index is calculated in real time.
Quantitative real-time PCR
Relative mRNA expression levels of target genes are measured by qPCR. Total RNA is extracted from cells or tissues with RNAiso Plus (TaKaRa, D9108A) following the manufacturer’s instructions, and cDNA is generated using a ReverTra Ace qPCR (Quantitative Real-time PCR) RT Kit (TOYOBO, FSQ-301). Real-time PCR is performed using SYBR Green PCR mix on a CFX ConnectTM Real-Time System normalized against GAPDH. The 2-ΔΔCt method is used to calculate relative gene expression levels. Gapdh is amplified as an internal control. Sequences of all primers for amplification of genes used in experiments are listed in Table S8.
Statistical analysis
After acquiring and storing the origin PA data, the median filtering algorithm and wavelet filtering algorithm are implemented based on a user-defined program for LabVIEW software. For each PA matrix, the various planes of the PA matrix are extracted. The horizontal plane and coronal plane of PA images are obtained. By applying for interpolated algorithm and reconstruction program, it is able to compute the MAP and depth-encoded by processing them through MATLAB program. The 3D image could be generated by arranging each matrix according to the time vector.
The MR images and the IF images are rendered with the software tool, RadiAnt, and ZEN, respectively. The merged images combing all the point-to-point positions of each pixel in a maximum-intensity projection fashion are fused using the software ImageJ, containing PA/PA image and IF/IF image. The ratio between the CVs signal area and the mLVs signal area is accessed to compare image features based on the different imaging methods. The mean PA intensity is calculated to reflect the condition of drainage and clearance. The mean PA intensity of a specific region is calculated, and the total value of the target region divided by the area of the target area can represent the average amount of tracer in the target region.
We estimated the local motion vectors from the consecutive frame and plotted their direction with color. According to the previous study, our system would capture the direction of lymph flow because the B-scan scanning velocity is less than the lymph velocity69,70,71,72. The mLVs that contained flowing tracers are first manually identified from the consecutive B-scan images. Run traces at points along these signals are then extracted. After thresholding the set of maximum amplitude, the images are fitted to estimate a flow direction.
Statistical analysis is conducted in SPSS software and GraphPad Prism 8. Data are expressed as the mean ± SEM. Significant differences are compared as noted in figure legends, using Student’s t test for statistical analysis in two-group comparison, and the differences are considered statistically significant at p < 0.05. Specific statistical parameters are detailed in the figure legends.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
Møllgård, K. et al. A mesothelium divides the subarachnoid space into functional compartments. Science 379, 84–88 (2023).
Fultz, N. E. et al. Coupled electrophysiological, hemodynamic, and cerebrospinal fluid oscillations in human sleep. Science 366, 628–631 (2019).
Alitalo, K. The lymphatic vasculature in disease. Nat. Med. 17, 1371–1380 (2011).
Wang, Y. D. & Oliver, G. Current views on the function of the lymphatic vasculature in health and disease. Genes Dev. 24, 2115–2126 (2010).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015).
Iliff, J. J. et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 4, 147ra111 (2012).
Drieu, A. et al. Parenchymal border macrophages regulate the flow dynamics of the cerebrospinal fluid. Nature 611, 585–593 (2022).
Semyachkina-Glushkovskaya, O. et al. Application of optical coherence tomography for in vivo monitoring of the meningeal lymphatic vessels during opening of blood-brain barrier: mechanisms of brain clearing. J. Biomed. Opt. 22, 121719 (2017).
Frederick, N. & Louveau, A. Meningeal lymphatics, immunity and neuroinflammation. Curr. Opin. Neurobiol. 62, 41–47 (2020).
Antila, S. et al. Development and plasticity of meningeal lymphatic vessels. J. Exp. Med. 214, 3645–3667 (2017).
Mestre, H., Mori, Y. & Nedergaard, M. The brain’s glymphatic system: current controversies. Trends Neurosci. 43, 458–466 (2020).
Louveau, A. et al. CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat. Neurosci. 21, 1380–1391 (2018).
Louveau, A. et al. Understanding the functions and relationships of the glymphatic system and meningeal lymphatics. J. Clin. Investig. 127, 3210–3219 (2017).
Ahn, J. H. et al. Meningeal lymphatic vessels at the skull base drain cerebrospinal fluid. Nature 572, 62–66 (2019).
Da Mesquita, S. et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature 560, 185–191 (2018).
Da Mesquita, S. et al. Meningeal lymphatics affect microglia responses and anti-Aβ immunotherapy. Nature 593, 255–260 (2021).
Shen, Q. et al. Gamma frequency light flicker regulates amyloid precursor protein trafficking for reducing β-amyloid load in Alzheimer’s disease model. Aging Cell 21, e13573 (2022).
Shen, Q. et al. Photobiomodulation suppresses JNK3 by activation of ERK/MKP7 to attenuate AMPA receptor endocytosis in Alzheimer’s disease. Aging Cell 20, e13289 (2021).
Ni, R. Q. et al. Multiscale optical and optoacoustic imaging of amyloid-β deposits in mice. Nat. Biomed. Eng. 6, 1031–1044 (2022).
Ding, X. B. et al. Impaired meningeal lymphatic drainage in patients with idiopathic Parkinson’s disease. Nat. Med. 27, 411–418 (2021).
Mestre, H. et al. Cerebrospinal fluid influx drives acute ischemic tissue swelling. Science 367, eaax7171 (2020).
Chen, J. M. et al. Meningeal lymphatics clear erythrocytes that arise from subarachnoid hemorrhage. Nat. Commun. 11, 3159 (2020).
Bolte, A. C. et al. Meningeal lymphatic dysfunction exacerbates traumatic brain injury pathogenesis. Nat. Commun. 11, 4524 (2020).
Li, X. J. et al. Meningeal lymphatic vessels mediate neurotropic viral drainage from the central nervous system. Nat. Neurosci. 25, 577–587 (2022).
Hsu, M. et al. Neuroinflammation creates an immune regulatory niche at the meningeal lymphatic vasculature near the cribriform plate. Nat. Immunol. 23, 581–593 (2022).
Hu, X. et al. Meningeal lymphatic vessels regulate brain tumor drainage and immunity. Cell Res. 30, 229–243 (2020).
Song, E. et al. VEGF-C-driven lymphatic drainage enables immunosurveillance of brain tumours. Nature 577, 689–694 (2020).
Dai, W. P. et al. A functional role of meningeal lymphatics in sex difference of stress susceptibility in mice. Nat. Commun. 13, 4825 (2022).
Ma, Q. L. et al. Outflow of cerebrospinal fluid is predominantly through lymphatic vessels and is reduced in aged mice. Nat. Commun. 8, 1434 (2017).
**e, L. L. et al. Sleep drives metabolite clearance from the adult brain. Science 342, 373–377 (2013).
Nowogrodzki, A. The world’s strongest MRI machines are pushing human imaging to new limits. Nature 563, 24–26 (2018).
Na, S. et al. Massively parallel functional photoacoustic computed tomography of the human brain. Nat. Biomed. Eng. 6, 584–592 (2022).
Absinta, M. et al. Human and nonhuman primate meninges harbor lymphatic vessels that can be visualized noninvasively by MRI. eLife 6, e29738 (2017).
Albayram, M. S. et al. Non-invasive MR imaging of human brain lymphatic networks with connections to cervical lymph nodes. Nat. Commun. 13, 203 (2022).
Kajita, H. et al. Visualization of lymphatic vessels using photoacoustic imaging. Keio J. Med. 70, 82–92 (2020).
Watanabe, S. et al. Photoacoustic lymphangiography is a possible alternative for lymphedema staging. J. Vasc. Surg.: Venous Lymphatic Disord. 10, 1318–1324.e2 (2022).
Suzuki, Y. et al. Subcutaneous lymphatic vessels in the lower extremities: comparison between photoacoustic lymphangiography and near-infrared fluorescence lymphangiography. Radiology 295, 469–474 (2020).
Pan, J. B. et al. Antigen-directed fabrication of a multifunctional nanovaccine with ultrahigh antigen loading efficiency for tumor photothermal-immunotherapy. Adv. Mater. 30, 1704408 (2018).
Laser Institute of America. ANSI Z136.1-2007 American National Standard for Safe Use of Lasers. (2007).
Wang, Z. Y. et al. Monitoring the perivascular cerebrospinal fluid dynamics of the glymphatic pathway using co-localized photoacoustic microscopy. Opt. Lett. 48, 2265–2268 (2023).
Wang, L. V. & Hu, S. Photoacoustic tomography: in vivo imaging from organelles to organs. Science 335, 1458–1462 (2012).
Taboada, C. et al. Glassfrogs conceal blood in their liver to maintain transparency. Science 378, 1315–1320 (2022).
Baik, J. W. et al. Super wide-field photoacoustic microscopy of animals and humans in vivo. IEEE Trans. Med. Imaging 39, 975–984 (2020).
Hindelang, B. et al. Enabling precision monitoring of psoriasis treatment by optoacoustic mesoscopy. Sci. Transl. Med. 14, eabm8059 (2022).
Liu, C. et al. Five-wavelength optical-resolution photoacoustic microscopy of blood and lymphatic vessels. Adv. Photonics 3, 016002 (2021).
Kim, J. et al. Super-resolution localization photoacoustic microscopy using intrinsic red blood cells as contrast absorbers. Light Sci. Appl. 8, 103 (2019).
Ahn, J. et al. In vivo photoacoustic monitoring of vasoconstriction induced by acute hyperglycemia. Photoacoustics 30, 100485 (2023).
Chen, J. B. et al. Confocal visible/NIR photoacoustic microscopy of tumors with structural, functional, and nanoprobe contrasts. Photonics Res. 8, 1875–1880 (2020).
Wang, Z. Y. et al. Photoacoustic-guided photothermal therapy by map** of tumor microvasculature and nanoparticle. Nanophotonics 10, 3359–3368 (2021).
Zhou, J. S. et al. Dual-modal imaging with non-contact photoacoustic microscopy and fluorescence microscopy. Opt. Lett. 46, 997–1000 (2021).
Pang, Z. et al. Multi-modality photoacoustic/ultrasound imaging based on a commercial ultrasound platform. Opt. Lett. 46, 4382–4385 (2021).
Zhang, W. et al. High-resolution, in vivo multimodal photoacoustic microscopy, optical coherence tomography, and fluorescence microscopy imaging of rabbit retinal neovascularization. Light Sci. Appl. 7, 103 (2018).
Liu, C. B. et al. The integrated high-resolution reflection-mode photoacoustic and fluorescence confocal microscopy. Photoacoustics 14, 12–18 (2019).
Wang, Z. Y. et al. Bifocal 532/1064 nm alternately illuminated photoacoustic microscopy for capturing deep vascular morphology in human skin. J. Eur. Acad. Dermatol. Venereol. 36, 51–59 (2022).
Lin, L. et al. High-speed three-dimensional photoacoustic computed tomography for preclinical research and clinical translation. Nat. Commun. 12, 882 (2021).
Yang, S. H. et al. Noninvasive monitoring of traumatic brain injury and post-traumatic rehabilitation with laser-induced photoacoustic imaging. Appl. Phys. Lett. 90, 243902 (2007).
Lv, J. et al. In vivo photoacoustic imaging dynamically monitors the structural and functional changes of ischemic stroke at a very early stage. Theranostics 10, 816–828 (2020).
Chen, Q. et al. Assessing hemorrhagic shock: feasibility of using an ultracompact photoacoustic microscope. J. Biophotonics 12, e201800348 (2019).
Deán-Ben, X. L. et al. Deep optoacoustic localization microangiography of ischemic stroke in mice. Nat. Commun. 14, 3584 (2023).
Li, L. et al. Single-impulse panoramic photoacoustic computed tomography of small-animal whole-body dynamics at high spatiotemporal resolution. Nat. Biomed. Eng. 1, 0071 (2017).
Chen, Z. Y. et al. Multimodal noninvasive functional neurophotonic imaging of murine brain-wide sensory responses. Adv. Sci. 9, 2105588 (2022).
**, T. et al. Photoacoustic imaging of brain functions: wide filed-of-view functional imaging with high spatiotemporal resolution. Laser Photonics Rev. 16, 2100304 (2022).
Wang, Z. Y. et al. Photoacoustic and ultrasound (PAUS) dermoscope with high sensitivity and penetration depth by using a bimorph transducer. J. Biophotonics 13, e202000145 (2020).
Zhang, D. C. et al. An ellipsoidal focused ultrasound transducer for extend-focus photoacoustic microscopy. IEEE Trans. Biomed. Eng. 68, 3748–3752 (2021).
Yang, F. et al. Wide-field monitoring and real-time local recording of microvascular networks on small animals with a dual-raster-scanned photoacoustic microscope. J. Biophotonics 13, e202000022 (2020).
Xavier, A. L. R. et al. Cannula implantation into the cisterna magna of rodents. J. Visualized Exp. 135, e57378 (2018).
Cheng, Z. W. et al. Subpixel and on-line motion correction for photoacoustic dermoscopy. IEEE J. Sel. Top. Quantum Electron. 27, 6800408 (2021).
Louveau, A. & Kipnis, J. Dissection and immunostaining of mouse whole-mount meninges. Protocol Exchange. https://doi.org/10.1038/protex.2015.047 (2015).
Takeda, K., Mori, S. & Kodama, T. Study of fluid dynamics reveals direct communications between lymphatic vessels and venous blood vessels at lymph nodes of mice. J. Immunol. Methods 445, 1–9 (2017).
Russo, E. et al. Intralymphatic CCL21 promotes tissue egress of dendritic cells through afferent lymphatic vessels. Cell Rep. 14, 1723–1734 (2016).
Kodama, T. et al. Visualization of fluid drainage pathways in lymphatic vessels and lymph nodes using a mouse model to test a lymphatic drug delivery system. Biomed. Opt. Expr. 6, 124–134 (2015).
Sharma, R. et al. Quantitative imaging of lymph function. Am. J. Physiol.-Heart Circulatory Physiol. 292, H3109–H3118 (2007).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (62335007; 62305118), the Natural Science Foundation of Guangdong Province (2022A1515010548), the Science and Technology Program of Guangzhou (2019050001), the Key Research and Development Plan of Guangzhou (202206010094), the STI2030-Major Projects (2022ZD0212200), the China Postdoctoral Science Foundation (2022M721223).
Author information
Authors and Affiliations
Contributions
S.Y. and F.Z. proposed the idea. F.Y. and Z.W. conceived the experiments and design the imaging system. F.Y., Z.W., and W.S. performed the experiments. X.L. provided the various probes used. R.M., E.W., W.X., and Z. Z. provided experimental assistance. Z.W., W.Z., S.Q., and M.W. provided useful advice. S.Y. and F.Z. supervised the overall projects. F.Y. wrote the initial draft of the manuscript. All authors discussed the results and contributed to manuscript writing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary information
41377_2024_1450_MOESM1_ESM.docx
Supplementary Information for: Ultra-high spatio-temporal resolution imaging with parallel acquisition-readout structured illumination microscopy (PAR-SIM)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, F., Wang, Z., Shi, W. et al. Advancing insights into in vivo meningeal lymphatic vessels with stereoscopic wide-field photoacoustic microscopy. Light Sci Appl 13, 96 (2024). https://doi.org/10.1038/s41377-024-01450-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41377-024-01450-0
- Springer Nature Limited