Abstract
Fusarium head blight (FHB) is a devastating disease lowering the yield and quality of wheat. Intensive use of limited allelic pools critically affects wheat resistance, while advancing pathogenic alleles. This study presents new alleles raised from moderate rather than elite crosses. ‘Parshall’ has indigenous moderate resistance that was crossed with the moderate susceptible ‘Reeder’ and developed the 110 RIL population (noted as PR) that was tested across three states and years in the USA, under open-field and greenhouse conditions. Heading- and FHB-related traits of incidence, severity, and index were assessed. A genetic map (1417 cM) of SNP/DArT markers was generated. Composite interval map** produced 45 QTL. Permutated LOD assessment identified two consistent QTL (4AL and 4BL) of type I and II resistance. Significant markers at the 4A QTL showed correlation with previous FHB resistance, while at the 4B QTL with pathogen responsive regions. The 1- and 2-D genome scans identified 16 QTL and six pairs of interacting markers for resistance, with additive effects between 0.17% and 2.19%; respectively. Additionally, additive × environment interaction was between 0.04% and 3.18% confirming the high environmental effect. Three pairs of QTL with additive × additive effects between 0.15% and 0.30% were revealed by FHB epistasis. However, the additive × additive environmental interaction at these three loci varied between 0.03% and 1%, confirming the G × E effect on FHB resistance. Hidden heritable epistasis was discovered in PR-population for FHB resistance. PR-alleles will help breeders develop lines mitigating epidemic environments and changeable climates, to boost yield facing hanger, food scarce, and secure worldwide penurious economies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Fusarium head blight (FHB) is caused by Fusarium graminearum Schwabe [teleomorph: Gibberella zeae Schw. (Petch)] [1]. Due to the asexual (conidia) and sexual (ascospores) life cycles of Fusarium, the infection can rapidly reach epidemic zenith [2].Once the pathogen lands on wheat spikes, FHB spores germinate a tube that enters plant cells through stomata and/or direct penetration [3]. Afterwards, FHB spreads within the apoplast causing cytological changes and cell death [4, 5]. At early infection, FHB reacts biotrophically and responsive resistance metabolites are released in tolerant cultivars [6]. Susceptible cultivars cause economic loss, where consequences exceed hundreds of millions of dollars a year in wheat growing regions of the US [7]. After the epidemics of 1990’s, wheat breeders around the world have united to defense the FHB raid. Based on various breeding methods, develo**/releasing resistant cultivars is established as the most efficient tool for mitigating FHB damages [8].
From the plant perspective, resistance against FHB is either morphological or physiological [9]. Avoidance mechanism, as a term, was used instead of morphological resistance. Short plants tend to be more susceptible to be infected relative to taller ones, probably due to the close distance of wheat heads to inoculum sources in soil [10]. Awned genotypes also showed higher infection compared to awnless ones, possibly due to increased surface area and more deposition of fungal spores. Avoidance traits were not proven to inhibit the pathogen after infection, however, they definitely enabled plants to escape the initial infection. While, physiological mechanisms of resistance involved biochemical pathways producing fungal inhibiting compounds [11]. Therefore, type I resistance is associated with morphological/avoidance mechanisms, where variation in such traits affects the number of infected heads [12]. From the pathogen perspective, resistance is composed of two models. The first model relates to pathogen biotrophic (within 6 h after infection (hai)) stage, where signals such as salicylic and phosphatidic acids, reactive oxygen species (ROS) and scavenging proteins are activated. Such activities are called plant hypersensitive responses and cell death, which help reduce pathogen spread. The second model relates to pathogen necrotrophic stage (between six and 24 hai), where ethanol signaling is activated to help transitioning from salicylic to jasmonic acids signaling [13] and slowing infection spread.
Many wheat cultivars are bred with prominent FHB resistance based on trusted germplasms such as Sumai3 (PI481542) and/or its derivatives [Ning7840 [14], CM82036 [15], DH181 [16], CJ9306 [17], and Line685 [18]). Genetically, FHB resistance is a complex quantitative trait that is classified into five discrete types [19]: (a) type I (resistance to initial infection denoted as incidence (INC)); (b) type II (resistance to pathogen spread denoted as severity (SEV)); (c) type III (resistance to accumulated FHB toxin levels denoted by deoxynivalenol (DON)), (d) type IV (resistance to FHB damaged kernel called (FDK)); and 5) type V (resistance through overall yield tolerance) [20]. Type II resistance assessment is the most reliable and least affected by environmental factors [21]. Generally, under controlled GH environments, type II resistance is the direct response of conidia injection and FHB spread [22]. On one level, some cultivars may demonstrate the resistance of type II based on one or more of the well-known QTL [5A/fhb5 [23], 3A [24], 3BS/fhb1 [25], 6B/fhb2 [26] and 2D [27]. Recently, glutathione S-transferase gene has naturally transferred from Epichloë (a widely distributed ascomycete fungal genus colonizing grasses) to the wild wheat grass (Thinopyrum ponticum) through a fungus-to-plant event that is denoted as Fhb7 QTL [28]. On another level, agronomic traits [plant height (HGT), spike-related traits, and heading dates (HD)] may have also an influence on FHB resistance of types I and II [29]. Indubitably, type II resistance is negatively associated with HGT, while positively associated with HD [30].
Originally, many QTL such as Fhb1 (3BS) were identified in the Chinese source Sumai3 [31]. Conditioned by the genetic background of wheat, Fhb1 locus was reported with 20–25% of FHB damage reduction [32]. It was found that pyramiding major QTL in a single cultivar boosts FHB resistance [33], while combining the effects of two QTL (Fhb1 and Fhb2) of Sumai3 may explain > 60% of PV. Later, an effective marker (umn10) was reported with close linkage to the Fhb1 QTL and widely used by many breeders [34]. The FHB biotrophic phase of pathogenicity delays the necrotrophic phase generating FHB resistance [5], where the environment limits both phases. Hence, the effects of alleles controlling FHB resistance have been dissected by both the high influence of environment and the complex nature of FHB resistance. Therefore, molecular-assisted breeding (MAB) can integrate multiple allelic effects and improve FHB resistance in wheat. However, MAB is considered successful if three factors are considered: (a) close linkage of marker/gene, (b) high consistency of validated genetics repeated across multiple environments/populations, and (c) use of cost- and time-effective methods for resistance assessment [35]. Consequently, the identification of new resistance alleles and generation of more genetic maps will enhance the applications of MAB in wheat. To better breed for FHB resistance (QTL), assessment under multiple environments is essential to discover alleles induced by the environment.
Intensive breeding, for Fhb1 and other QTL of type II resistance, is slowing the progress of building resistance beyond the levels of parents [36]. Moderate resistance is attributed to single QTL existence, while enhancement of resistance is to pyramiding multiple QTL in a genotype. For example, building type II and III (DON) resistance has enhanced phenotypic variation (PV) by 93 and 56%, respectively [37]. Except for Fhb1, QTL explains less than 20% of PV [29]. From the biochemical resistance perspective, upon infection, some defense genes are induced providing variable responses [13]. Such responses may prevent initial infection (type I) and slow the spread of fungus (type II) [38, 39]. Pathogenesis-related proteins are induced as well, upon infection [40]. Proteomic and transcriptomic studies revealed 163 genes and 37 proteins to be induced by FHB infection [14].
A population of 110 RILF14 (referred as PR) was developed from the cross between Parshall (PI-613587) and Reeder (PI-613586) (http://wheatpedigree.net/). Parshall has moderate resistance to FHB, while Reeder has moderate susceptibility. Both Parshall and Reeder have medium early heading stage and a common parent (ND-674). Pedigree, field/GH trials, and biochemical quality tests have all confirmed that the source of FHB resistance in ‘Parshall’ is domestic and non-Sumai3 [41]. Thus come the objectives of current study as to: (a) genetically produce a map/identify QTL for FHB resistance in PR population, (b) provide breeders with the stable and consistent QTL/alleles/markers for FHB resistance, (c) offer correlation among alleles of HD and those of FHB resistance, (d) deliver epistatic/pleotropic/interaction effects among FHB associated loci, and (e) enrich the allelic variation in the genetic pool of wheat, where QTL for FHB resistance are essentially sought/applied by breeders worldwide.
2 Materials and methods
2.1 Plant materials and field experiments/FHB inoculation
Both parents (Parshall and Reeder) and the PR population were developed by the breeding program at the North Dakota State University (NDSU) following the standard guidelines of growing wheat plants. The PR population (RIL-derived-F14) is evaluated under field and GH experiments for two seasons at 5 geographical locations that all administered by the universities contributed through the authors to this study. Briefly, one location each in Minnesota (MN) (Minneapolis, 44°59′N, 93°16′W) and South Dakota (SD) (44°19′05″N, 96°47′00″W), and three locations in North Dakota (ND) [Carrington (47°45′00″N, 99°12′39″W), Prosper (46°96′30″N, 97°01′98″W) and Langdon (48°45′42″N, 98°22′18″W)]. All experiments were conducted as randomized complete block design (RCBD) with four replicates in ND and two replicates in MN and SD. Three checks were evaluated in all experiments, locations, and across all years including highly resistant checks (Alsen ‘PI-615543’ and ND2710 ‘PI-633976’) and highly susceptible check (ND2398). Field experiments in ND were conducted using FHB mist-irrigation nurseries method [9] and grain spawn as an artificial inoculation [42]. The experimental unit consisted of hill/plot of 0.3 m length. Each hill/plot is phenotypically assessed and averaged using at least 10 plants. FHB inoculum was applied in the field at flowering time and FHB-related traits were evaluated three weeks after inoculation.
2.2 Greenhouse experiments and FHB inoculation
All RIL, parents and checks were planted in the GH and laid out in an RCBD with four replicates using an eight-inch-sized pots. The planting soil was Sunshine Mix#1 (Sun Gro Horticulture, Agawam, MA USA), which was mixed with 20 g of Osmocote® slow release fertilizer (Scott’s Company LLC, Marysville, OH, USA). Each pot contained five plants. At flowering time, five spikes of each pot were artificially infected using FHB spore-suspension method, i.e. direct injection of inoculum in a single middle spikelet [43, http://www.triticate.com.au) for (DArT) genoty** [2.5 Constructing the genetic map of PR population Out of genotypic markers, only polymorphic DArT (559) and SNP (441) markers were used and converted based on parental codes. Deviation of markers data from expected ratio (1:1) for RIL was assessed using χ2 test and distorted markers were deleted and 513 markers (244 DArT and 269 SNP) were resulted. Linkage groups (LG) were constructed using CarthaGène [46], where maximum distance of 30 cM and a minimum LOD threshold of four were used. Co-segregating markers are merged into single markers. Most probable positions of markers along the LG are determined using the commands [mrkdouble, mrkmerges, group 0.3, 4 mrkselset, build, flips, polish, detail]. Kosambi map** function is set [47]. Each LG is compared to the wheat consensus SSR and AFLP genetic maps [48] available in GrainGenes database (http://wheat.pw.usda.gov/ggpages/maps.shtml). Final maps were validated based on the DArT [49] and SNP consensus maps using AutoGRAPH software [50] (http://autograph.genouest.org/). Composite interval map** (CIM) method [51] was performed using QGene 4.0 [52]. A scanning interval of 1 cM and a window size of 5 cM were used to detect QTL. QTL were referred in the text to either single locus or multiple loci. The number of markers controlling the background was set to five markers at forward and revers regression. A QTL was considered significant at LOD = 2.5. Permutation test was set at 1000 replications with 30 threads to confirm LOD score threshold at each single trait across all markers. The total number of assessed environments for INC, SEV, NDX, and HD were five, 10, seven, and six; receptively, then confirmed by the combined mean across assigned environments of each trait (AI, AS, GH, AN, and HD). Confidence interval (CI) of flanking markers (FM) was appointed at the two positions of LOD = 2 flanking each peak of a single QTL. The proportion of PV explained by each single QTL was determined by the square of correlation coefficient (R2). QTL with overlap** CIs were considered as one QTL for each traits (Table 6) and even if repeated in different traits (Table 5). QTL explaining ≥ 10% of PV was considered major [53], and if expressed across ≥ 50% of tested environments were considered stable/consistent. Additive QTL effects had positive and negative signs that indicate the contribution of Parshall and Reeder toward higher trait mean values; respectively. The size of additive effect contributed by QTL for each resistance type was divided into different sets following [54]. Graphical illustration of LG and QTL was presented using MapChart 2.2 software [55]. Windows QTL Cartographer V2.5 [56] and QTL Network 2.0 were used to analyze allelic effects of main additive (a), epistatic (aa) and interaction of QTL across all environments (ae and aae) [57]. A 1000 permutation test was used to calculate critical F-values for an experiment-wise significance level of 0.05. Tests to detect QTL interaction were conducted at 1 cM intervals with a window size of 5 cM, where backward elimination of non-significant spurious QTL was performed [58]. Since FHB-related traits reflect the pathogenic symptoms, high mean values of a trait (SEV, INC, NDX) reflect low resistance. For HD, the higher the mean value the lateness of the genotype. Analysis of variance (ANOVA) was performed using SAS 9.3® (SAS Institute, Cary, NC), where genotypes (G) were considered as having fixed effect, while other sources of variation as having random effect. For each individual environment, data was analyzed as RCBD. The statistical model of F-max ratio for homogeneity [54] was performed to test the possibility to combine environments and significance of G, E and G × E interactions. Briefly, if the F-max value of the ratio between major and minor experimental errors of tested environments was ≤ 10 then combining the means was considered possible. ANOVA was conducted using Proc Mixed model for individual and combined environments (AI, AS, GH, AN, and HD) of each trait. Pearson’s coefficient of correlations (r) was estimated using Proc Corr of the SAS 9.3 between the means of combined environment at each traits. Significance of Pearsons’s correlations, F-test, and ANOVA was set at p ≤ 0.05 value. Means separation of genotypes was determined using F-protected least significant differences (LSD, p ≤ 0.05) from the Proc Mixed model analysis. Broad-sense heritability (H2) was calculated based on family means as described by [59] using the random ANOVA Proc Mixed model output, excluding the means of parents and checks. The equation used to calculate heritability was: H2 = σ2G/[ σ2G + ( σ2GE/e) + ( σ2E/re)], where σ2G = genotypic variance, σ2GE = genotype × environment interaction, σ2E = experimental error variance, e = total number of environments, and r = total number of replications within an environment.2.6 QTL map** and interaction among FHB-related traits
2.7 Statistical analysis of phenotypic data
3 Results
3.1 Variation and correlation among FHB-related traits
Analysis of variance for INC, SEV (A), and HD (ND) resulted in highly significant effects of all sources of variance (Table 1). Likewise, a high significance was observed for NDX trait along with HD and other FHB-associated traits (INC and SEV) probably due to the G × E interaction. These data confirm the high influence of environment on FHB traits. The mean squares of genotypes were larger than G × E interaction resulting in high broad-sense heritability (H2 = 0.52 and 0.79) for ASEV (A) of combined field data and HD (ND). In GH-SEV data, significant effects of G and Rep/E resulted, however, in moderate heritability (H2 = 0.32). Higher heritability (H2 = 0.45) was found for NDX, while the lowest heritability (H2 = 0.16) was expressed for INC. Pearson correlation coefficient between combined environments for HD and for FHB-related traits were presented in Table 2. A-SEV data was highly significant and positively correlated (r = + 0.10, + 0.37***, + 0.86***, + 0.34***, and + 0.39*** at P ≤ 0.001) with GHSEV, A-INC, A-NDX, ND-HD, and MN10-HD traits (Table 2).
The values of means for parents, checks and RIL, LSD, minimum and maximum means for FHB-related and HD traits were presented in Table 3. The mean values of assessed traits in PR population showed a normal distribution curve (Fig. 1). All phenotypic means [INC (A), SEV (AS and G), and NDX (A, S12, and P12)] for Parshall (the moderate resistant parent) were lower (91.3, 26.5, 53.7, 25.1, 30.3, and 40.1%) compared to that of Reeder (the moderate susceptible parent) (95.1, 34.4, 65.8, 25.9, 30.5, and 48.6%). Despite these low values (resistance), Parshall did not express the expected differential values compared to Reeder. This has raised the question about the exact source of enhanced (transgressive) resistance for FHB in several PR lines. On one hand, many RIL have showed transgressive higher mean values (100, 37.8, 75.8, 36.9, 41.3, 64.1%) exceeding the value of the moderate susceptible parent (Reeder) for FHB-related traits and therefore classified as susceptible. On the other hand, other RIL have showed lower mean values (30.2, 21.2, 24.7, 16.9, 17.0, 14.8%) than the moderate resistant parent (Parshall) and therefore classified as resistant lines, similar or even superior than the two most resistant checks Alsen and ND2710 (Table 3).
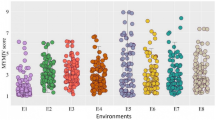
Frequency means distribution of PR population for FHB-related traits (SEV, INC, and NDX) and HD. Combined means across environments was the one used for all distributions unless noted to specify the environment. Arrows indicate the means of the two parents (Parshall and Reeder). A Frequency distribution of PR population for incidence trait across all open-field environments; B Frequency distribution for severity trait across all open-filed environments; C Frequency distribution for severity across all GH environments; D Frequency distribution of index across all environments except “Prosper and South Dakota”; E Frequency distribution for index in single environment “Prosper” in 2012; F Frequency distribution for index across South Dakota environments in 2012; G Frequency distribution for heading date across all environments of North Dakota; H Frequency distribution for heading date across all environments in Minnesota
3.2 Linkage map and QTL Analysis For PR population
Out of the 513 polymorphic DArT and SNP markers, 501 markers were mapped to 28 linkage groups (LG) for 19 chromosomes with 278 unique loci (Table 4). Our PR mapped loci look to be more accurate than the maps of [60, 61] of 624 markers (189 DArT) on 21 LG. The LG were assigned individually to the specified wheat chromosomes based on the corresponding markers anchored from the consensus SNP and DArT maps, then compared to maps of GrainGenes (https://wheat.pw.usda.gov/GG3/maps-short). A total of 10, 11 and seven LG were assigned to the A, B, and D genomes; respectively. However, no markers were assigned to 1D and 4D chromosomes. The total distance of the map was 1417 cM, where 745.7, 581.1 and 90.2 cM were; respectively, assigned to the A, B, and D genomes. The current map distance is close to one third of the wheat genetic map, which is ≥ 10 cM limit suitable for QTL scanning [62]. The average distance between two markers was 5.1 cM. Consensus maps published by [43, 63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78] were all used to validate identified QTL in current study (Fig. 2 and 3). A total of 12 QTL were consistent and associated with multiple traits for FHB resistance and tagged-initial as Qfhb in Table 5. Among these, 11 QTL have shown major effects (≥ 10% of PV). Five of seven QTL were associated with HD with major effect. As four major QTL (Qfhb.ndsu-3B2, Qfhb.ndsu-4A2, Qfhb.ndsu-4B2, and Qfhb.ndsu-5B) were associated with FHB traits and HD across four to 10 environments (Table 5), five HD QTL were detected consistently in two to five environments.
QTL detected with epistatic effects for FHB-related traits and HD in PR population. Linkage group names are assigned at the top of each chromosome. QTL intervals were shown in bars, while the position of effective markers were shown by arrow-head. QTL for FHB-related traits including INC is shown in blue, SEV is shown in red, and NDX is shown in yellow, while the HD is shown in green. QTL with interacting markers for epistatic effects were linked by black bars ended with a colored circular tip corresponding to the color of major trait associated with these interacting markers/QTL
3.2.1 QTL for enhanced FHB resistance of type I in PR population
For type I resistance, a total of five QTL were detected on five different chromosomes (Qinc.ndsu-4A2, Qinc.ndsu-4B2, Qinc.ndsu-5B, Qinc.ndsu-6D, and Qinc.ndsu-7A). These QTL were identified using the means of five environments (M10, S10, C11, P11, and P12) and confirmed by the combined mean (AI) across all environments (Table 5, Fig. 2). Except for the Qinc.ndsu-6D (LOD = 2.6), all identified QTL for type I resistance had significant LOD score (2.9 to 4.5). Meanwhile three QTL (Qinc.ndsu-4A2, Qinc.ndsu-4B2, and Qinc.ndsu-7A) showed relatively higher PV (15.56%, 10.02%, and 10.01%; respectively).
QTL allocated on PR linkage groups validated across consensus maps of wheat for FHB-related traits and HD. Outer circle represents the linkage group of PR population. Inner circle represents the consensus and corresponding segments of each linkage group. Black threads represents the common markers detected in both maps. Panel A, the genetic map of the consensus linkage group of markers found in genome A; Panel B, the genetic map of the consensus linkage group of markers found in genome B. Panel C, the genetic map of the consensus linkage group of markers found in genome D. QTL intervals were presented as bars inside the center of circles. The graph is drawn using R and MS-PowerPoint based on presented epistatic loci in Table 9
Based on the classifications of allelic effective size [79], three sets of QTL with low allelic additive effects were detected in this study as indicators for enhanced FHB resistance. These QTL were contributed by the moderate susceptible parent (Reeder) affecting the mean values of INC trait. In the first set, one QTL (Qinc.ndsu-7A) had allelic additive effect (-1.25). that was detected under two environments (P10 and P11) with maximum PV of 10.01%. Meanwhile, a QTL (Qinc.ndsu-4B2) was attributed to Reeder and detected under a wider range of environments (P10, M10, P11, L11, S11, C11, P12, S12, AI, AN, and AS) across three states (ND, SD, and MN), with major additive effects (-2.89) and high PV (10.02%) (Table 6). In the meanwhile, the observed allelic additive effects from Reeder were detected across 10 environments. The third set of lowest additive effect (3.7) was attributed to Parshall at the Qinc.ndsu-4A2, which explained 15.56% of INC PV. It was also consistent across nine environments in the three states (ND, SD, and MN). Overall, the total pyramided additive effect, contributed by the alleles from Reeder at three loci on the LGs 4B2, 5B, and 7A for resistance type I, was 5.41 vs 6.14 from Parshall at 2 different loci on the genomes 4A and 6D. This has resulted in lowering mean values of INC and increasing resistance of type I. Two major and consistent QTL for resistance type I, one was provided by Reeder at Qinc.ndsu-4B2 and the other was provided by Parshall at Qinc.ndsu-4A2. This hints contribution of B-genome by Reeder and A-genome from Parshall for FHB resistance. Also, it suggests recombination events at B-genome by Reeder and at A-genome by Parshall. It emphasizes the epistatic events observed between A and B genomes (Figs. 2 and 4).
3.2.2 QTL for enhanced FHB resistance of type II in PR population
For resistance type II, 16 QTL (Qsev.ndsu-1A2, Qsev.ndsu-2A2, Qsev.ndsu-3A.a, Qsev.ndsu-3A.b, Qsev.ndsu-3A.c, Qsev.ndsu-3A.d, Qsev.ndsu-4A2, Qsev.ndsu-4B2, Qsev.ndsu-5A, Qsev.ndsu-6B.a, Qsev.ndsu-6B.b, Qsev.ndsu-6B.c, Qsev.ndsu-6D, Qsev.ndsu-7A.a, Qsev.ndsu-7A.b, and Qsev.ndsu-7B) were located on 11 chromosomes (Table 6, Fig. 2). These QTL were identified based on data of 10 environments (P10, M10, S10, C11, P11, S11 G11, P12, S12, and G12). All QTL identified for resistance type II had major effects (R2 ≥ 10%) and significant LOD scores (3.2 – 7.9). The allelic effects from Reeder ranged from -1.76 to -6.83 and contributed to increasing the mean values (susceptibility) at six QTL (Qsev.ndsu-3A.a, Qsev.ndsu-3A.c, Qsev.ndsu-3A.d, Qsev.ndsu-6B.c, Qsev.ndsu-7A.b, and Qsev.ndsu-7B). Likewise, the allelic additive effects from Parshall ranged from 1.36 to 6.08 that increased the mean values at the other nine QTL (Qsev.ndsu-1A2, Qsev.ndsu-2A2, Qsev.ndsu-3A.b, Qsev.ndsu-4B2, Qsev.ndsu-5A, Qsev.ndsu-6B.a, Qsev.ndsu-6B.b, Qsev.ndsu-6D, and Qsev.ndsu-7A.a). Overall, the total pyramided allelic additive effects of Parshall (31.5%) were less than of Reeder (-32.49%) toward increasing mean values of severity (susceptibility).
Therefore, based on the size of inherited parental additive effects, the type II resistance in the PR population can be divided into three sets [79]: (a) the set of QTL with additive effects less than 2, (b) the set of QTL with additive effects between 2 and ±3.47, and (c) the set of QTL with additive effects higher than ±3.47). The first set includes five QTL (Qsev.ndsu-2A2, Qsev.ndsu-5A, Qsev.ndsu-6B.a, Qsev.ndsu-6B.b, and Qsev.ndsu-7Ab) with additive effects equal to 1.36, 1.62, 1.61, 1.81, and -1.76; respectively. These QTL had low effects toward increasing mean values of severity (resistance). Noticeably, these five QTL are expressed under SD environment with major effects explaining PV values between 11.26% and 14.80%. The accumulated additive effects contributed by Parshall was 6.4 (at four QTL) vs only 1.76 (at one QTL) from Reeder. Four out of the five QTL showed association with FHB-index and one QTL with HD.
The second set included four QTL (Qsev.ndsu-4A2, Qsev.ndsu-6B.c, Qsev.ndsu-6D, and Qsev.ndsu-7A.a) with additive effects equal to 3.47, −2.82, 3.3, and 3.37; respectively. These QTL have moderate effects on increasing values of severity and consequently moderate resistance. These four QTL were expressed well under ND environments with major effects explaining between 10.68 and 16.58% of the PV. The accumulated additive effects contributed by Parshall was 10.14 vs only −8.98 from Reeder. Two (Qsev.ndsu-6D and Qsev.ndsu-7A.a) out of the four QTL were contributed by Parshall with major effects explaining 10.68% and 11.47% of PV; respectively, under ND environments. Likewise, one QTL (Qsev.ndsu-6B.c) out of the four QTL were contributed by Reeder with major effects explaining 16.58% of PV under ND environments. Lastly, the Qsev.ndsu-4A2 QTL of the four was attributed by both Parshall and Reeder with major effects explaining 15.25% of PV across the environments of the three states ND, SD and MN.
The third set included seven QTL (Qsev.ndsu-1A2, Qsev.ndsu-3A.a, Qsev.ndsu-3A.b, Qsev.ndsu-3A.c, Qsev.ndsu-3A.d, Qsev.ndsu-4B2, and Qsev.ndsu-7B) with additive effects of 6.08, −4.16, 4.39, −4.88, −585, 4.49, and −6.83; respectively. These QTL had high effect toward increasing the values of severity and therefore, FHB susceptibility. Six out of these seven QTL were more effective under GH. Only one QTL (Qsev.ndsu-4B2) had major and consistent effect under multiple environments across ND, SD, and MN explaining 25.03% of PV. Three (Qsev.ndsu-1A2, Qsev.ndsu-3A.b, and Qsev.ndsu-4B2) out of the seven QTL contributed by Parshall with major effect explaining 28.70, 19.71, and 25.03% of PV; respectively. Four (Qsev.ndsu-3A.a, Qsev.ndsu-3A.c, Qsev.ndsu-3A.d, and Qsev.ndsu-7B) out of the seven QTL have major effects explaining 19, 12.62, 22.54, and 24.04% of PV; respectively and contributed by Reeder under GH. The total additive effects contributed by the moderate resistant parent Parshall was 14.96 compared to 21.75 from the moderate susceptible parent Reeder.
3.2.3 QTL for FHB-index in PR population
For FHB-index, CIM analysis resulted in 10 QTL (Qndx.ndsu-1A2, Qndx.ndsu-3A, Qndx.ndsu-3B2, Qndx.ndsu-4A2, Qndx.ndsu-4B2, Qndx.ndsu-5A, Qndx.ndsu-6B.a, Qndx.ndsu-6B.b, Qndx.ndsu-6B.c, and Qndx.ndsu-7A.b) on eight chromosomes (Table 6, Fig. 2). These QTL were identified using the means of seven datasets (M10, S10, C11, P11, S11, P12, and S12) and confirmed by overall mean of combined environments (AN). All QTL identified for FHB-index had major effects (R2 ≥ 10%) and highly significant LOD (scores ranging between 3.1 and 7.3). Identified QTL for FHB-index can be divided into three sets based on the size of additive effects [79] contributed by the parents as follow: (a) the set of QTL with additive effects less than 2, (b) the set of QTL with additive effects between 2 and 3.57, and (c) the set of QTL with additive effects higher than 3.57.
The first set included four QTL (Qndx.ndsu-3B2, Qndx.ndsu-5A, Qndx.ndsu-6B.b, and Qndx.ndsu-7A.b) with additive effects equal to 1.21, 1.62, 1.81, and -1.76; respectively. Three out of these QTL were contributed by Parshall under GH environments and expressed major effects explaining 10.98, 12.38, and 14.80% of PV; respectively. Only Qndx.ndsu-3B2 QTL was consistent across four environments, mainly under GH environments. Another QTL (Qndx.ndsu-7A.b) was expressed with major effect explaining 14.90% of PV and contributed by Reeder under one environment. As expected, the total accumulated allelic effects inherited from the moderate resistant parent Parshall was as high as 4.64 vs only -1.76 from the moderate susceptible parent Reeder showing the resistance acquired.
The second set included three QTL (Qndx.ndsu-3A, Qndx.ndsu-6B.a and Qndx.ndsu-6B.c) with additive effects of 2.59, 3.57 and -2.91; respectively. Two out of the three QTL were located on 3A and 6B.a LG and sourced from Parshall with major effects explaining 13.18 and 17.47% of PV; respectively. Another QTL on 6B.c LG was originated from Reeder with major effects explaining 16.68 of PV. Overall, the total additive effects attributed to Parshall was 6.16 compared to 5.61 from Reeder showing the moderate resistance acquired.
The third set included three QTL (Qndx.ndsu-1A2, Qndx.ndsu-4A2, and Qndx.ndsu-4B2) with additive effects equal to 6.08, 3.58, and 4.44; respectively. All the three QTL were from Parshall that explained 28.70, 18.74, and 14.22% of PV; respectively. Two of the three QTL on 4A2 and 4B2 LG were consistent and effective under at least nine environments across the three states ND, SD, and MN. Therefore, these two QTL are of great importance. The overall additive effects for resistance to FHB-index contributed by Reeder (-7.37%) were less than those of Parshall (24.9%) showing the important role of Parshall.
3.2.4 QTL for heading date in PR population
For HD, eight QTL (Qhd.ndsu-2A2.a, Qhd.ndsu-2A2.b, Qhd.ndsu-4A2, Qhd.ndsu-4B1, Qhd.ndsu-4B2, Qhd.ndsu-5A.a, Qhd.ndsu-6B, and Qhd.ndsu-7A.b) were identified on eight chromosomes (Table 6, Fig. 2). These QTL were identified using the means of six environments (M10, P10, C11, L11, P11, and P12) and confirmed by the overall mean of five combined environments (AI, AS, AN, ND, and GH). Eight QTL (Qhd.ndsu-2A2.a, Qhd.ndsu-2A2.b, Qhd.ndsu-4A2, Qhd.ndsu-4B1, Qhd.ndsu-4B2, Qhd.ndsu-5A.a, Qhd.ndsu-6B, and Qhd.ndsu-7A.b) for HD showed significant LOD score (between 2.8 and 6.5) with wide range of major effects (R2 = 10.47–52.85%). The allelic additive effects of Reeder ranged between −0.52 and −0.79% toward increasing the mean values of HD (lateness) at two QTL (Qhd.ndsu-4B1 and Qhd.ndsu-5A.b). Similarly, the allelic additive effects of Parshall ranged between 0.56 and 2.01% toward earliness at the remained seven QTL (Qhd.ndsu-2A2.a, Qhd.ndsu-2A2.b, Qhd.ndsu-4A2, Qhd.ndsu-4B2, Qhd.ndsu-5A.a, Qhd.ndsu-6B, and Qhd.ndsu-7A.b). The total pyramided allelic additive effects (−1.71%) due to the alleles of Reeder is less than those of Parshall (9.39%). The QTL identified for HD can be divided into two sets based on the size of additive effects [79] a set of QTL with additive effects less than 2, and (b) a set of QTL with additive effects above 2.
The first set included seven QTL (Qhd.ndsu-2A2.a, Qhd.ndsu-2A2.b, Qhd.ndsu-4B1, Qhd.ndsu-4B2, Qhd.ndsu-5A.a, Qhd.ndsu-6B, and Qhd.ndsu-7A.b) with additive effects equal to 0.96, 0.56, −0.52, 0.89, 1.46, 1.11, and 0.9; respectively. Six out of these QTL were contributed by Parshall and encoded for earliness (0.96, 0.56, 0.89, 1.46, 1.11, and 0.9) with major effects explaining 22.32, 11.35, 13.12, 29.5, 17.52, and 12.35%; of PV; respectively. The other QTL was from Reeder and contributed to lateness (−0.52) with major effect explaining 10.47% of PV. The total pyramided allelic additive effects (−1.71%) were due to the alleles from Reeder and less than those of Parshall (9.39%). In addition, the two QTL 4A and 4B had important alleles, mainly from Parshall, with significant accumulated effects. Since both parents had medium earliness, this shows a correlation between alleles of resistance and those for HD. QTL expression under one (Qhd.ndsu-5A.a and Qhd.ndsu-7A.b), two (Qhd.ndsu-2A2.a), three (Qhd.ndsu-2A2.b and Qhd.ndsu-4B1), five (Qhd.ndsu-6B), and 10 (Qhd.ndsu-4B2) environments for HD confirmed the quantitative nature of HD and the diversity of allelic effects observed in PR. The second set included a single consistent QTL (Qhd.ndsu-4A2) for HD from Parshall that was expressed in nine environments with the largest effect (2.01) explaining 53% of PV.
3.3 Pleiotropy and association effects among QTL
Six FHB-related QTL with allelic markers on three LGs (4A2, 6B, and 3A) were interacting pleiotropically with markers on the other three LGs (4B2, 7A, and 3B1) for FHB-index and HD (Table 7 and Figs. 2 and 4). The flanking markers (WSP7442-WSP7077) on the centric intervals (48.9–52.9 cM) for FHB-index on the 4A2 LG were associated with those (WPt8892-WPt6149) on the distal intervals (6.9–9.3 cM) of the short arm of 4B2 LG for the same trait. The remaining five pairs of interacting QTL were reported for HD. The flanking markers (WSP921-WPt7203) on the centric interval (49.3–55.9 cM) of the 6B LG for HD were associated with the markers (WPt7763-WPt0551) on the distal interval (130.2–130.7 cM) of the long arm of 7A LG for HD. Similarly, the flanking markers (WPt7203-WPt5673) on the long arm interval (55.9–63.0 cM) of the 6B LG for HD were associated with the same markers (WPt7763-WPt0551) on the distal interval (130.2–130.7 cM) on the long arm of 7A LG. Likewise, the flanking markers (WPt5673-WPt6585) on the long arm position (63.0 cM) of the 6B LG for HD were associated with the markers (WPt2260-WPt7763) on the distal interval (108.7–130.2 cM) of the long arm of LG 7A (Tables 6 and 7). The same flanking markers (WPt5673-WPt6585) on the distal position (63.0 cM) of 6B LG were associated with the flanking markers (WPt7763-WPt0551) on the distal intervals (130.2–130.7 cM) of the long arm of 7A LG for HD. The last pair of interacting QTL for HD was detected for the flanking markers (WSP4053-WSP1673) at the distal intervals (120–121.8 cM) of the 3A LG that were associated with the flanking markers (WPt8238-WSP280) on the distal interval (64.8–69.9 cM) of the long arm 3B1 LG.
Epistatic interacting QTL mapped on PR linkage groups for FHB-related traits and HD. Blue bold left-sided ribbons represents the linkage groups of B genome of PR population. The gray right-sided ribbons represents the linkage groups of A genome of PR population. Epistatic and interacting markers were connected by ribbons inside the circle. Flanking regions of each QTL were detailed in Table 9
3.4 Heritability and interaction among FHB associated QTL
Seven QTL for SEV, NDX, and HD were mapped to five LGs (2A2, 4A2, 4B2, 5B, and 6B) with significant additive effects. These QTL explained between 0.17 and 2.19% of PV produced by the value of broad sense heritability (H2(a)) with additive effects (a1) ranging from 0.51 to 1.80% (Tables 7 and 8). The heritability (as one component of genetic gain) due to environmental preference at Qsev.ndsu-4A2.b QTL was high (H2 (ae) = 1.03%), where Reeder has contributed alleles at three environments aE2, aE3, and aE7. For NDX trait, two QTL on 4A2 and 4B2 were identified. The Qndx.ndsu-4A2 QTL at the markers interval (48.9–52.9 cM) was having high and significant additive effect (a1 = 0.95***) with heritability H2 (a) = 1.29%. This QTL has also expressed significant additive × environment effect at aE2, aE3, aE6, and aE7 with high heritability as well (H2 (ae) = 3.18%). For this QTL, Parshall has contributed effects at a1, aE2, aE3, and aE5, while Reeder has contributed effects at aE6 and aE7 only. The second NDX QTL was Qndx.ndsu-4B2 with significant additive × environment effect (a1 = 1.15***) that explained heritability (H2 (a) = 2.19). At this QTL, Reeder has contributed allelic effects at four environments aE2, aE3, aE6, and aE7. While, Parshall has contributed effects at three environments a1, aE4, and aE5. For HD, four QTL on 2A2, 4B1, 5B, and 6B LG were identified (Table 8).
3.5 Heritability, epistasis and environmental interaction of FHB associated QTL
Herein, the markers (WSP7442-WSP7077) at the intervals (48.9–52.9 cM) on 4A2 QTL showed epistatic effect with the markers (WPt8892-WPt6149) at the intervals (6.9–9.3 cM) related to 4B2 QTL (Table 9). The additive × additive effect due to these two QTL was significant with heritable effect at each QTL [80] as H2 (aa) = 0.15% (Table 9, Fig. 4). The heritability portion is due to interaction with the environment at each QTL (H2 (aae) = 1%). The markers (WSP4053-WSP1673) related to 3A QTL at the intervals 120–121.8 cM were epistatically interacting with the markers (WPt8238-WSP280) linked to 3B1 QTL at the interval 64.8–69.9 cM. This interaction resulted in significant effects for Reeder with heritability of (aa) 0.25% and of (aae) 0.06. The markers (WPt5673-WPt6585) of the second QTL (Qhd.ndsu-6B) for HD were epistatically interacting as well with the markers (WPt7763-WPt0551) on 7A QTL at the interval 130.2–130.7 cM. Parshall has dominated the allelic effects at these two interacting loci with aa1 = 2.62 and aaE2 = 0.06. The heritability of the additive (aa) effect was 0.3% and for additive × epistasis (aae) was = 0.03%. Despite the low values of additive × epistasis (aae) effects (0.03 and 0.06), its detectable existence gives value to the significance of the G × E effect on FHB resistance in wheat. Besides, it is the epistatic direction towards enhancing the genetic gain of the trait that matters, not only the value of the effect these quantitative loci carry. The more number of loci pyramided together (even with low effects), the higher the influence of these loci on a trait, which is the power behind quantitative genetics of a trait like FHB resistance. This assumes that the hidden epistasis for FHB might be due to its low quantitative effect values.
4 Discussion
4.1 Improving FHB resistance in PR RIL population
In the current study, the RIL showed moderate enhancement for type I resistance (Table 1). This is most likely due to the alleles from the moderate resistant parent (Parshall). Compared to the mid-parent value (30.45%), type II resistance of RIL was expressed as average reaction (29.70%) between the moderate resistant parent (Parshall—26.50%) and the moderate susceptible parent (Reeder—34.40%). Also, these values were comparable to those expressed by the resistant checks Alsen (26.40%) and ND2710 (26.70%). Notably for RIL and parents, FHB severity in the field and GH (G) environments, were having moderate values comparable to that of the resistant checks [Alsen (37.40%) and ND2710 (44.10%)] and much less than the susceptible check [ND2398 (68.30%)]. The average value of RIL showed higher resistance of type II (51%) than of both moderate parents (Parshall for resistance—53.70% and Reeder for susceptibility—65.80%). This is a clear indication that both parents contributed alleles of type II resistance to the RIL. These results are supported by previous studies on assessing the effectiveness of specific combining ability in F2 populations under GH conditions [81] using direct inoculation method under GH conditions [82] or when using grain-spawn inoculation methods used under field conditions [83].
Similarly, the FHB-index, under field environments, of parents (Parshall 25.10% and Reeder 25.9%) was in average between the resistant checks [Alsen (20%) and ND2710 (26.8%)] and the highly susceptible check [ND2398 (60.50%)]. Fortunately, the value of FHB-Index for the RIL population was also average (27%) dragged towards resistance not susceptibility of the check’s value (60.5%). This implies that most resistant alleles of RIL were possibly affected by the moderate resistant parent (Parshall ) compared to the susceptible parent (Reeder ). Besides, the close values of FHB-index detected in parents (Parshall 25.10% and Reeder 25.9%) could be sourced from the common ancestor (ND-674). Results of NDX, INC, and SEV for RIL and its parents, have highlighted several points. First, RIL exposed higher values (susceptibility) for INC (93%) under field environments; while revealed lower values for resistance as NDX and SEV (27.4 and 29,5%; respectively, Table 1); and showed moderate SEV values under GH environments. This suggests that RIL showed overall higher resistance under field environments, while total moderate resistance under GH environments. This agrees with previous reports underlining the complexity of FHB resistance and the high effects of environments on FHB infection [12]. It also confirms previous report that spike inoculation is an accurate and direct assessment method for FHB type II resistance [21]. Finally, population structure, as previously reported to affect FHB resistance [32, 35], pyramiding alleles from moderate resistant parents can directly contribute to various levels of resistance in a population compared to the alleles from highly resistant parents. This is also confirmed by previous studies where about 1604 wheat lines were generated from crosses between 120 females and 15 males and found to meet the variety-type employed to improve FHB resistance [84]. Consequently, both moderate parents in PR population contributed to enhance FHB resistance, where alleles of resistance type I are discrete from those of type II.
Under ND and MN environments, Parshall was slightly earlier (0.2–0.5 day) than Reeder (Table 3). However, RIL population in average (56 days), was relatively two days later than parents (54 days). Such deviation could be due to both parents lateness alleles specifically Reeder’s. Nevertheless, some RIL showed transgressive behavior towards earliness (52.8 days). Though, alleles from both parents resulted in slight late heading (0.5 day) under ND compared to MN environment. This slight difference in HD between both parents suggests their adaptation to the US North Central Plains, the quantitative nature, and the high impact of environments on the INC, SEV, NDX and HD traits [23,24,25,26,27,28,29].
The value of heritability for SEV was relatively higher than INC in our study. Meanwhile, type II resistance under GH conditions was negatively correlated (Table 3) with type I and HD but was positively correlated with FHB-index under field. This suggests the significant impact of environment on these traits that most likely controlled by many genes with minor effects. HD was positively correlated with all FHB-related traits under field conditions, while negatively correlated under GH environments displaying significant role of HD on FHB expression and its resistance as well demonstrated in previous studies (Table 3, [8, 85,86,88,89].
4.1.1 Susceptible parent can enhance FHB type I in PR population
In this study, we demonstrated that among the five QTL that were identified for type I resistance based on allelic additive effects (Table 5), two QTL Qinc.ndsu-5B and Qinc.ndsu-7A were contributed by Reeder, the moderate susceptible parent. Similar results were reported previously in a study using 218 RIL, where the moderate susceptible parent Alondra in the cross Ning894037/Alondra contributed some enhancement to FHB resistance [90]. However, the FHB resistance contributed by Parshall at the Qinc.ndsu-6D QTL was expressed exclusively under ND environments, while the effects of Qinc.ndsu-4B2 QTL contributed by Reeder were detected across the three states, ND, SD, and MN. Similarly, the enhanced effect of the Qinc.ndsu-4A2 from Parshall was major and consistent across the three states of ND, SD, and MN as well. This proves the stability of resistance transferred to PR population from Parshall. This confirms similar effects observed for the resistant parent Ernie in a study by [91]. The total additive effects contributed by Reeder alleles at three loci on the LGs 4B2, 5B, and 7A, were higher than that from Parshall at the 4A2 and 6D LGs for type I resistance suggesting that the inherited type I resistance in PR population probably originated from Reeder. In addition, the enhancement for type I resistance expressed in PR population could not be due to allelic additive effects inherited from one parent only but most likely from a combination from both parents as was the QTL at the 4B2 LG from Reeder and at the 4A2 LG from Parshall for type I resistance. Parshall type I resistance response was reflected by a higher pyramided additive allelic effects limiting early biotrophic FHB infection. This reaction could be the reason explaining the purple color seen on the infected spikes of Parshall. In a study by [92] a purple acid phosphatase was reported in some resistant lines. However, the purple color requires further metabolic study in Parshall to confirm if the color was exclusively a symptomatic reaction to the high phenolic compounds slowing down FHB spread along the spike.
4.1.2 Parshall sourced enhancement for FHB Type II resistance in PR population
CIM analysis for type II resistance revealed 17 QTL dispersed across 11 different LGs (Table 6, Figs. 2 and 3). The allelic effects of Reeder on FHB type II resistance were attributed to seven QTL, while allelic effects of Parshall were attributed to nine QTL. Parshall had less allelic effects (31.5%) than Reeder (-32.49%) toward increasing mean values of FHB severity. Since Parshall is the moderate resistant parent, then alleles from Parshall may had dominant resistance effects and/or less additive effects towards increasing FHB severity (susceptibility) in PR population. Therefore, as expected, the contribution for FHB type II resistance was mostly from the indigenous-moderate resistant parent (Parshall). Our findings supports previous studies using moderate resistant and indigenous wheat genotypes as an alternative short-term strategy of QTL incorporation [60, 93, 94]. Therefore, type II resistance in PR population was expressed by the five QTL on the LGs of 2A2, 5A, 6B.a, 6B.b and 7Ab. Among these, four QTL were from Parshall and one from Reeder supporting the above hypothesis. The second category included five QTL on the LGs of 4A2, 5B, 6Bc, 6D, and 7A.a with moderate effects to increasing FHB severity and consequently moderate resistance. The contribution of Parshall was overall higher compared to Reeder. Under ND environments, two QTL on the LGs of 6D and 7A.a out of the five were from Parshall and two QTL on the LGs of 5B and 6B.c were from Reeder. The fifth QTL on 4A LG was a co-contribution from both parents and effective across three states ND, SD and MN environments. Similar results combining moderate effects from both parents to enhance FHB resistance in PR population was also reported by previous studies [61, 95].
4.1.3 Parshall enhanced FHB-index resistance in PR population
FHB-index showed three categories within 11 QTL on eight chromosomes (Table 6, Figs. 2 and 3) with major effects and significance (LOD ≅ 3.1–7.3). The first category included four QTL on the LGs of 3B2, 5A, 6B.b, and 7A.b with more resistance alleles from Parshall (three out of the four QTL) and had major effects, while the fourth QTL was form Reeder. Accordingly, the accumulated allelic effects of resistance were inherited from Parshall more (4.64) than Reeder (1.76). The second category included another four QTL on the LGs of 3A, 6B.a, 6B.c, and 7A.a with moderate resistance, where the QTL on the LGs of 3A and 6B.a were major and contributed by Parshall and the other two QTL on the LGs of 6B.c and 7A.a were from Reeder. For this category, the accumulated additive effects from Parshall (6.16) were also higher than Reeder (5.61). The last category was solely from Parshall with three QTL on the LGs of 1A2, 4A2, and 4B2, where the QTL on the LGs of 4A2 and 4B2 were consistent across the three states ND, SD, and MN. Interestingly, the detected alleles of moderate resistance from Parshall (24.9%) were more frequent than the moderate susceptibility from Reeder (-7.37%). Hence, combining more genes/QTL of FHB resistance will be more effective to breeders than relying on single gene effect such as Fhb1 [36]. Overall, most FHB QTL explain < 20% of PV (except for Fhb1) [29]. This study presents many QTL with ≥ 20% of PV (Table 5 and Table 6) providing breeders with abundant alternatives other than Fhb1 towards acceptable levels of FHB resistance. Such QTL would not be discovered if was analysed based on elite genetics of crossing parents. This proves the strength of moderate genetic analysis.
4.2 Importance of major and consistent QTL on 4A and 4B chromosomes
Type I resistance is, generally, either morphological or physiological [9]. Morphologically, taller genotypes showed lower severity relative to short ones, most likely due to the distance between spikes and inoculum sources on the soil [10]. Similarly, awnless genotypes showed lower infection compared to awned probably due to reduced surface area and less deposition of fungal spores. Such resistance can be attributed to avoidance mechanism that allow plants to escape initial infection. Therefore, type I resistance is mainly associated with avoidance mechanisms hence typically affects the number of infected heads [12]. The type II and I resistance (avoidance) in PR population was associated with the stable Qfhb.ndsu-4A2 and Qfhb.ndsu-4B2 QTL that were contributed by both parents across nine to 10 environments (Table 5). Their stability can increase the chances of general combining ability (GCA) of FHB resistance as reported by [96], where GCA of FHB was attributed to the additive effects of parental genotypes in an F2 winter triticale populations, while specific combining ability (SCA) could be attributed to the non-additive allelic interaction. For FHB-index and HD, the 4A2 and 4B2 QTL have explained major additive allelic effects from both parents. Previous studies have also reported interesting results for FHB resistance in the same region on 4AS (33.51–58.31 cM) of our study. Using a population of 188 RIL from the cross BR18/BRS179, a QTL in the same region (6.6–42.20 cM) has explained 10.1% PV of FHB [95]. Similar results were reported by using a population of 282 RIL of the cross Ningmai9/Yangmai158, where the same region on 4A (32.57–34.58 cM) has explained 5.48% PV of FHB resistance [97]. Likewise, a study using 212 RIL population from the Cutler/AC-Barrie cross, has identified a region (37.63–37.77 cM) on 4A that explained 10.68% PV of FHB resistance [98]. Finally, the region 14.46–40.85 cM was associated with resistance of type II in the wheat consensus map between the two markers IWB70645 and IWB36120 [99]. Based on above studies, the 4A2S QTL, as well in our study, was consistent across multiple populations and environments; emphasizes its importance to FHB resistance. Our finding is also supported by [100], who indicated that genes contributed by susceptible parents could enhance the genetics of resistance to FHB. Finally, our results based on an arguably smaller number of RIL population (110 RIL), was not a factor halting the identification of such important QTL like that on 4A [101].
Our 4BS region (0.0–21.10 cM) for FHB resistance in PR population was associated with resistance to various diseases as the region (12.00–16.00 cM) has explained 20.7% of PV for Stagonospora glume blotch disease in a RIL population of the cross Arina/Forno [102]. A neighboring 4BS region (6.90–7.50 cM) was linked to glume blotch resistance in doubled-haploid lines generated from the cross 6HRWSN125/WAWHT2074 that explained 4% of the PV [103]. In another RIL population of the cross Florett/Biscay, a 4BS region (2.00–10.00 cM) has explained 9.3% of PV for Septoria tritici resistance [104]. Similarly, the region (0.00–10.00 cM) on 4BS explained 18.3% PV of Karnal bunt resistance in a RIL population of the cross WL711/HD29 [105]. However, the region (3.76–6.18 cM) on 4BS between wPt9824 and wPt1272 markers, was associated with FHB type III resistance [99], compared consensus maps for three traits (plant height, spike length, and thousand-kernel weight) in three RIL populations and found that the 4BS region was associated with a QTL for many traits including a QTL for plant height (QPH.caas-4BS) and two spike length QTL (qSL.4B-1 and qSL.4B-2) [65]. Another QTL for spike length was denoted as qSL-4B-1 on 4BS and a QTL (qTKW-4B-1) for TKW at 4 cM position. These studies emphasize the importance of the alleles on 4BS region in our PR population for diseases resistance in general, and for FHB in particular. This also emphasizes the quality of using our PR to identify the intervals on 4BS with only 110 RIL, a result that was supported by many studies using larger population sizes. The only debatable note is the wider intervals identified for PR QTL, if compared to other intervals identified by larger population sizes in previous studies. However, for plant height and spike length traits, the intervals in big-sized population [65] was noted as wide (00–26.5 cM and 6.94–34.98 cM) similar to our findings. Existence of morphological alleles (plant height and spike length) along with physiological alleles (multiple disease resistance), supports the proposed avoidance resistance placed in QTL such as 4BS in PR population.
4.3 Pleotropic effects of FHB and non-FHB alleles in PR population
The importance of FHB loci in PR population was highlighted by a number of associated traits at the same loci (Table 5). For example, the intervals harboring Qfhb.ndsu-1A2 was previously reported for plant height at qPH-1A (11.8 cM) [106]. Another QTL (qTKW-2A-1 (0.2 cM)) for TKW was associated with Qhd.ndsu-2A2 QTL in our study [71]. The chromosomal region of Qfhb.ndsu-3A in our study was reported for plant height at qPH-3A-1 (34.20 cM and 41.93 cM) and at QPH.caas.3AS (45.85 cM), and for spike length at qSL-3A-2 (0.0–28.6 cM) [65]. The interval positions of PR (Qfhb.ndsu-3B2) QTL was previously reported by [71] for plant height at qPH-3B-1 (3.40–11.00 cM) and spike length at qSL-3B-1 (1 cM). The location of PR Qhd.ndsu-5A.a and -5A.b QTL were previously reported for plant height at qPH-5A-1 (37.3 cM and 76.40–86.20 cM) and at QPH.caas.5AS (37.02 cM), and for TKW at qTKW-5A-1 (65.4–84.6 cM). Likewise, the positions of PR Qfhb.ndsu-6B.b and Qfhb.ndsu-6B.c QTL were reported for TKW at qTKW-6B-1 (37.2 cM) and qTKW-6B-2 (64.4 cM); respectively. The PR QTL Qsev.ndsu-6B.b was once reported for plant height in a multiple-QTL model study at 37.1–38.7 cM. Similarly, the PR Qndx.ndsu-7A.a QTL was associated with spike length at qSL-7A-1 (24 cM) and also for relative maturity at 60.3–61.9 cM and 46.9–47.6 cM (Table 6). This combined effects of alleles for INC and maturity at this location supports the relationship between the alleles for FHB and HD at the 4B2 and 4A2 QTL of PR population. Therefore, pleotropic effects of many FHB QTL identified in our study (Table 9) have previously been confirmed by many studies. Similarly, 12 QTL were reported at many intervals on 1A including 1–4 cM, 4–6 cM, 7–9 cM, 0–1 cM, and 6–7 cM for INC, SEV, NDX and maturity in the cross Blackbird/Strong-Field, and coincided with the intervals of our Qsev.ndsu-1A2 QTL in PR. Additionally, a QTL (Qsev.ndsu-3A.b) in PR was reported twice for FHB resistance at two intervals (62–69 cM and 54.1–55.1 cM), and another QTL (Qsev.ndsu-3A.c) was associated with plant height at the interval 75.2–81.9 cM in the cross Blackbird/Strong-Field. These results confirmed the role of the pleiotropic effects, of many FHB resistance alleles in PR, on several agronomic, resistance to many diseases and yield traits as well as to SCA of hidden non-additive allelic interactions [95].
4.4 Effect of population background on FHB reaction
Breeding history of FHB resistance in wheat underline the contribution of germplasm traced back to Asia, including China with Sumai3 that recognized as a major source of resistance [107]. However, the agronomic performance of Sumai3 varies widely due to its non-adaptation (undesirable linkages) to many environments outside of its original environment. Therefore, finding other sources of well-adapted FHB resistance as alternative germlines to Sumai3, even if these alternatives were less resistant, is paramount [108]. Parshall, a native, adapted and with moderate resistance has been used successfully in ND breeding programs, and herein, as a parent to replace susceptible alleles from the well adapted cultivar Reeder. Breeding programs worldwide used intensively the derivatives of Sumai3 for this purpose. However, many successful wheat breeding programs did not rely on Sumai3 as a source of resistance due to many “bad” linkages such as yield drag for instance. This was evidenced by the panel of resistant spring wheat lines used in a genome wide association study by [109]. In this study, 14 lines (non-Sumai3) were identified with FHB resistance under different environments. Additionally, wheat breeding programs at CIMMYT have developed and distributed lines to other countries that resulted in develo** many elite FHB resistance genotypes that were non-related to Sumai3 [110].
The fact that resistance in Parshall is a non-Sumai3 makes Parshall a good alternative genetic pool for FHB resistance in wheat breeding programs. For decades, studying FHB resistance in wheat depended on populations developed by crosses between highly resistant and highly susceptible parents [111]. Only few studies used moderate resistant or susceptible parental sources in their breeding method to develop population for FHB selection. This includes Ernie [112], Freedom [113], and Roane [114]. Based on our results, where PR population derived from the cross between the moderate resistant (Parshall) and the moderate susceptible (Reeder), FHB resistance alleles can be pyramided from both parents. Therefore, these two parents can be added to the list of parents used to improve many traits related to FHB resistance as well as other agronomic traits through pleiotropic effects for instance. This supports the proposition that susceptible effects of some alleles do not necessarily render the targeted resistance [115]. However, negative effects of some susceptible alleles could contribute positively to improve some other quantitative traits or even other resistances to other diseases [116, 117]. In fact, our study showed that FHB susceptibility alleles provided by Reeder in the PR population had some positive effects toward enhancing resistance such as the alleles in 4B2 QTL for type I and in 4A2 for type II FHB resistances.
5 Conclusion
Ecological instability, currently known as climate change, adds pressure on plant breeders to develop new cultivars with high adaptability and resistance to the ever-changing pathogens. For more than three decades, wheat breeding for FHB resistance relied intensively on narrow genetic source (Chinese sources such as Sumai3 and its derivatives). This dependency has created a limited allelic variation in wheat resistance, while allowed for pathogen to develop spores with high severity alleles combined with favorable environmental conditions to spread. Therefore, wheat breeders are forced to continually identify new alleles of resistance, dig inside exotic wheat materials and/or search for endogenous and hidden alleles of resistance in well-adapted germplasms. Our study has provided PR population as an input for all above targets for breeders. Briefly first, new alleles of resistance laid in 45 QTL, where 20 QTL of them were related to multiple traits and stable across multiple environments and years. Such alleles are of great importance and wide applications in wheat breeding due to their high adaptability to the Northern Plain environments. Second, the allelic diversity in the type of resistance (five QTL for type I and 17 QTL for type II) provides breeding programs with stable alleles of FHB resistance under multiple environments. Third, Parshall exhibited a major impact on several alleles of FHB resistance types. Identified alleles of QTL for type I resistance have limited the pathogen to infect plants, while other alleles of QTL for type II resistance have slowed the spread of the disease. Along with Parshall alleles, PR acquired unique combination of allelic effects for resistance of type II from Reeder, the alleged susceptible parent to FHB. Despite its size (110 RIL), PR population produced a unique combination of hidden pleiotropic resistance would not be discovered unless the epistatic QTL alleles (such as Qhd.ndsu-3A and -6B) were introduced through medium genetics of resistance in both parents. Additionally, concurrency of PR QTL positions, effects, functions, and associated traits with those of previous reports, depleted any critic about the population size effect on the efficiency of QTL identification. Fourth, allelic effects identified for FHB resistance in PR, sourced from either Parshall, Reeder, or both were verified across multiple environments and years. Finally, the study will be of great help to wheat breeders for mitigating the negative impacts of climate change favoring FHB pathogens.
Data availability
“The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.”
References
McMullen M, Jones R, Gallenberg D. Scab of wheat and barley: A re-emerging disease of devastating impact. Plant Dis. 1997;81:1340–8.
Gilbert J, Fernando W. Epidemiology and biological control of Gibberella zeae/Fusarium graminearum. Canad J Plant Pathol. 2004;26:464–72.
Seong KY, Zhao X, Xu J, Güldener U, Kistler HC. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet Biol. 2008;45:389–99. https://doi.org/10.1016/j.fgb.2007.10.008.
Gunnaiah R, Kushalappa AC, Duggavathi R, Fox S, Somers DJ. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE. 2012;7: e40695. https://doi.org/10.1371/journal.pone.0040695.
Zhuang Y, Gala A, Yen Y. Identification of functional genic components of major Fusarium head blight resistance quantitative trait loci in wheat cultivar Sumai3. Mol Plant Microbe Interact. 2013;26:442–50. https://doi.org/10.1094/MPMI-09-12-0217-R.
Basnet B, Glover K, Ibrahim A, Yen Y, Chao S. A QTL on chromosome 2DS of ‘Sumai 3’ increases susceptibility to Fusarium head blight in wheat. Euphyt. 2012;186:91–101. https://doi.org/10.1007/s10681-012-0709-1.
Nganje WE, Bangsund DA, Leistritz FL, Wilson WW, Tiapo NM. Regional economic impacts of Fusarium Head blight in wheat and barley. Rev Agri Econ. 2011;26(3):332–47. https://doi.org/10.1111/j.1467-9353.2005.00228.x.
Lu Q, Lillemo M, Skinnes H, He X, Shi J, Ji F, Dong Y, Bjørnstad A. Anther extrusion and plant height are associated with Type I resistance to Fusarium head blight in bread wheat line ‘Shanghai-3/Catbird.’ Theor Appl Genet. 2013. https://doi.org/10.1007/s00122-012-1981-9.
Rudd JC, Horsley RD, McKendry AL, Elias EM. Host plant resistance genes for Fusarium head blight: sources mechanisms and utility in conventional breeding systems. Crop Sci. 2001;41:620–7.
Mesterhazy A. Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 1995;114:377–86.
Gilsinger J, Kong L, Shen X, Ohm H. DNA markers associated with low Fusarium head blight incidence and narrow flower opening in wheat. Theor Appl Genet. 2005;110:1218–25. https://doi.org/10.1007/s00122-005-1969-8.
Saharan MS. Current status of resistant source to Fusarium head blight disease of wheat: a review. Indi Phytopath. 2020;73:3–9. https://doi.org/10.1007/s42360-019-00186-x.
Ding L, Xu H, Yi H, Yang L, Kong Z, Zhang L, Ma Z. Resistance to hemi-biotrophic F graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS ONE. 2011;6(4): e19008. https://doi.org/10.1371/journal.pone.0019008.
Zhou W, Kolb F, Bai G, Shaner G, Domier L. Genetic analysis of scab resistance QTL in wheat with microsatellite and AFLP markers. Geno. 2002;45:719–27. https://doi.org/10.1006/geno.2002.6780.
Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierschneider M, Ruckenbauer P. Molecular map** of QTLs for Fusarium head blight resistance in spring wheat I Resistance to fungal spread (Type II resistance). Theor Appl Genet. 2002;104:84–91. https://doi.org/10.1007/s001220200009.
Yang ZP, Gibert J, Fedak G, Somers D. Genetic characterization of QTL associated with resistance to Fusarium head blight in a doubled-haploid spring wheat population. Geno. 2005;48:187–96. https://doi.org/10.1016/j.ygeno.2005.03.001.
Jiang GL, Dong Y, Shi J, Ward RW. QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306 II Resistance to deoxynivalenol accumulation and grain yield loss. Theor Appl Genet. 2007;115:1043–52. https://doi.org/10.1007/s00122-007-0623-2.
Lu QX, Szabo-Hever A, Bjørnstad A, Lillemo M, Semagn K, Mesterhazy A. Two major resistances QTL were required to counteract the increased susceptibility to Fusarium head blight at the Rht-D1b gene in wheat. Crop Sci. 2011;51:2430–8. https://doi.org/10.2135/cropsci2011.02.0084.
Mesterházy Á, Bartók T, Mirocha CG, Komoróczy R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999;118:97–110. https://doi.org/10.1046/j.1439-0523.1999.118002097.x.
Terzi V, Tumino G, Stanca AM, Morcia C. Review: reducing the incident of cereal head blight and mycotoxins in small grain cereals in Europe. J Cereal Sci. 2014;59:284–93. https://doi.org/10.1016/j.jcs.2013.12.005.
Bai GH, Shaner G. Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 1996;80:975–9.
Buerstmayr M, Alimari A, Steiner B, Buerstmayr H. Genetic map** of QTL for resistance to Fusarium head blight spread (type 2 resistance) in a Triticum dicoccoides x Triticum durum backcross-derived population. Theor Appl Genet. 2013;126:2825–34. https://doi.org/10.1007/s00122-013-2165-9.
Xue S, Xu F, Tang M, Zhou Y, Li G, An X, et al. Precise map** Fhb5 a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L). Theor Appl Genet. 2011;123:1055–63. https://doi.org/10.1007/s00122-011-1652-7.
Yu JB, Bai GH, Zhou WC, Dong YH, Kolb FL. Quantitative trait loci for Fusarium head blight resistance in a recombinant inbred population of Wangshuibai/Wheaton. Phytopath. 2008;98:87–94. https://doi.org/10.1094/PHYTO-98-1-0087.
Anderson JA, Stack RW, Liu S, Waldron BL, Fjeld AD, et al. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor Appl Genet. 2001;102:1164–8. https://doi.org/10.1007/s001220000509.
Cuthbert P, Somers D, Brule´-Babel A. Map** of Fhb2 on chromosome 6BS: a gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L). Theor Appl Genet. 2007;114:429–37. https://doi.org/10.1007/s00122-006-0439-3.
Lin F, Xue SL, Zhang ZZ, Zhang CQ, Kong ZX, Yao GQ, Tian DG, et al. Map** QTL associated with resistance to Fusarium head blight in the Nanda2419×Wangshuibai population II: Type I resistance. Theor Appl Genet. 2006;112:528–35. https://doi.org/10.1007/s00122-005-0149-8.
Wang H, Sun S, Ge W, Zhao L, Hou B, Wang K, et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:5435. https://doi.org/10.1126/science.aba5435.
Buerstmayr H, Ban T, Anderson JA. QTL map** and marker-assisted selection for Fusarium head blight resistance in wheat; a review. Plant Breed. 2009;128:1–26. https://doi.org/10.1111/j.1439-0523.2008.01550.x.
Lu Q, Lillemo M, Skinnes H, He X, Shi J, Ji F, et al. Anther extrusion and plant height are associated with type I resistance to Fusarium head blight in bread wheat line “Shanghai-3/Catbird.” Theor Appl Genet. 2012;126:317–34. https://doi.org/10.1007/s00122-012-1981-9.
Waldron BL, Moreno-Sevilla B, Anderson JA, Stack RW, Frohberg RC. RFLP map** of QTL for Fusarium head blight resistance in wheat. Crop Sci. 1999;39:805–11. https://doi.org/10.2135/cropsci1999.0011183X003900030032x.
Pumphrey MO, Bernardo R, Anderson JA. Validating the Fhb1 QTL for Fusarium head blight resistance in near isogenic wheat lines developed from breeding populations. Crop Sci. 2007;47:200–6. https://doi.org/10.2135/cropsci2006.03.0181.
Miedaner T, Wilde F, Steiner B, Buerstmayr H, Korzun V, et al. Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxynivalenol (DON) content and disease severity. Crop Sci. 2006;46:2393–400. https://doi.org/10.2135/cropsci2006.03.0182.
Liu S, Pumphrey MO, Gill BS, Trick HN, Zhang JX, Dolezel J, et al. Toward positional cloning of Fhb1 a major QTL for Fusarium head blight resistance in wheat. Cere Res Commun. 2008;36:195–201.
Randhawa HS, Asif M, Pozniak C, Clarke JM, Graf RJ, Fox SL, et al. Application of molecular markers to wheat breeding in Canada. Plant Breed. 2013;132:458–71. https://doi.org/10.1111/pbr.12051.
- Mesterhazy A, Buerstmayr H, Toth B, Lehoczki-Krsjak Sz, SzaboHever Á, Lemmens M. An improved strategy for breeding FHB resistant wheat must include Type I resistance. In: Proc of the 5th Canadian Workshop on Fusarium Head Blight. Delta Winnipeg. 2007. pp. 51–66.
Lemmens M, Scholz U, Berthiller F, Dall’Asta C, Koutnik A, Schuhmacher R, et al. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol Plant Micro Interact. 2005;18:1318–24. https://doi.org/10.1094/MPMI-18-1318.
Bowles DJ. Defense-related proteins in higher plants. Annu Rev Biochem. 1990;59:873–907. https://doi.org/10.1146/annurev.bi.59.070190.004301.
Pritsch C, Muehlbauer GJ, Bushnell WR, Somers DA, Vance CP. Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol Plant Microb Interact. 2000;13:159–69. https://doi.org/10.1094/MPMI.2000.13.2.159.
Mergoum M, Frohberg RC, Stack RW, Olson T, Friesen TL, Rasmussen JB. Registration of ‘Glenn’ wheat. Crop Sci. 2006;46:473–4. https://doi.org/10.2135/cropsci2005.07-0223.
Stack RW, Frohberg RC. Fusarium head blight of spring wheat in an inoculated and irrigated nursery 1996. Biol Cult Test Cont Plant Dis. 1997;12:111.
- Bekele GT. Head SCAB screening methods used in CIMMYT. In: Wheats for more tropical environments. CIMMYT Mexico D F Mexico. 1995. pp. 169–173.
ElDoliefy AEF, Kumar A, Anderson JA, Glover KD, Mamidi S, Elias EM, et al. Genetic dissection of Fusarium head blight resistance in spring wheat cv ‘Glenn.’ Euphyt. 2020;216:71. https://doi.org/10.1007/s10681-020-02610-0.
Akbari M, Wenzl P, Caig V, Carling J, **a L, Yang SY, et al. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet. 2006;113:1409–20. https://doi.org/10.1007/s00122-006-0365-4.
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–51. https://doi.org/10.1038/nbt.2969.
De Givry S, Bouchez M, Chabrier P, Milan D, Schiex T. CarthaGene: multi-population integrated genetic and radiation hybrid map**. Bioinfo. 2005;21:1703–4. https://doi.org/10.1093/bioinformatics/bti222.
Kosambi DD. The estimation of map distances from recombinant values. Ann Eugen. 1944;12:172–5.
Somers D, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet. 2004;109:1105–14. https://doi.org/10.1007/s00122-004-1740-7.
Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J. 2012;10:826–39. https://doi.org/10.1111/j.1467-7652.2012.00702.x.
Derrien T, Andre C, Galibert F, Hitte C. AutoGRAPH: an interactive web server for automating and visualizing comparative genome maps. Bioinfo. 2007;23:498–9. https://doi.org/10.1093/bioinformatics/btl618.
Zeng ZB. Precision map** of quantitative trait loci. Genet. 1994;136:1457–68.
Joehanes R, Nelson JC. QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics. 2008;24:2788–9. https://doi.org/10.1093/bioinformatics/btn516.
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK. An introduction to markers, quantitative trait loci (QTL) map** and marker-assisted selection for crop improvement: the basic concepts. Euphyt. 2005;142:169–96. https://doi.org/10.1007/s10681-005-1681-5.
Leppink J, O’Sullivan P, Winston K. Effect size–large, medium, and small. Perspect Med Educ. 2016;5(6):347–9. https://doi.org/10.1007/s40037-016-0308-y.
Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–8. https://doi.org/10.1093/jhered/93.1.77.
Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. 2012. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Yang J, Hu CC, Hu H, Yu RD, ** and visualizing genetic architecture of complex traits in experimental populations. Bioinfo. 2008;24:721–3. https://doi.org/10.1093/bioinformatics/btm494.
Laurie CC, Wang S, Carlini-Garcia LA, Zeng ZB. Map** epistatic quantitative trait loci. BMC Genet. 2014;15:112. https://doi.org/10.1186/1471-2156-15-112.
Tabachnick BG, Fidell LS. Using multivariate statistics. Boston, MA: Allyn and Bacon Inc; 2001.
Holland JB, Nyquist WE, Cervantes-Martínez CT. Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev. 2003;22:9–111.
Semagn K, Bjørnstad Å, Skinnes H, Marøy AG, Tarkegne Y, William M. Distribution of DArT, AFLP, and SSR markers in a genetic linkage map of a double haploid hexaploid wheat population. Geno. 2006;49:545–55. https://doi.org/10.1139/G06-002.
Semagn K, Skinnes H, Bjørnstad Å, Marøy AG, Tarkegne Y. Quantitative trait loci controlling Fusarium head blight resistance and low deoxynivalenol content in hexaploid wheat population from ‘Arina’ to NK93604. Crop Sci. 2007;47:294–303. https://doi.org/10.2135/cropsci2006.02.0095.
Doerge RW. Multifactorial genetics: map** and analysis of quantitative trait loci in experimental populations. Nat Rev Genet. 2002;3:43–52. https://doi.org/10.1038/nrg703.
Rocher F, Alouane T, Philippe G, Martin ML, Label P, Langin T, et al. Fusarium graminearum infection strategy in wheat involves a highly conserved genetic program that controls the expression of a core effectome. Int J Mol Sci. 2022;23:1914. https://doi.org/10.3390/ijms23031914.
Qu P, Wang J, Wen W, Gao F, Liu J, ** of wheat (Triticum aestivum L) using 90K SNP array. Front Plant Sci. 2021;12: 727077. https://doi.org/10.3389/fpls.2021.727077.
Hu X, Yingquan Z, **gjuan Z, Shahidul I, Maoyun S, Yun Z, et al. Consensus genetic linkage map construction based on one common parental line for QTL map** in wheat. Agro. 2021;11(2):227. https://doi.org/10.3390/agronomy11020227.
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, et al. Multiple wheat genomes reveal global variation in modern breeding. Nat. 2020;588:277–83.
Venske E, Dos Santos RS, Farias DDR, Rother V, da Maia LC, Pegoraro C, et al. Meta-analysis of the QTLome of Fusarium head blight resistance in bread wheat: refining the current puzzle. Front Plant Sci. 2019;10:727.
Cai J, Wang S, Su Z, Li T, Zhang X. Meta-analysis of QTL for Fusarium head blight resistance in Chinese wheat landraces. The Crop J. 2019. https://doi.org/10.1016/j.cj.2019.05.003.
Hu X, Rocheleau H, McCartney C, et al. Identification and map** of expressed genes associated with the 2DL QTL for Fusarium head blight resistance in the wheat line Wuhan 1. BMC Genet. 2019;20:47. https://doi.org/10.1186/s12863-019-0748-6.
Sari E, Berraies S, Knox RE, Singh AK, Ruan Y, Cuthbert RD, et al. High density genetic map** of Fusarium head blight resistance QTL in tetraploid wheat. PLoS ONE. 2018;13(10): e0204362. https://doi.org/10.1371/journal.pone.0204362.
Wen W, He Z, Gao F, Liu J, ** H, Zhai S, et al. A high-density consensus map of common wheat integrating four map** populations scanned by the 90K SNP array. Front Plant Sci. 2017;8:1389.
Clavijo BJ, Venturini L, Schudoma C, et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Gen Res. 2017;27(5):885–96. https://doi.org/10.1101/gr.217117.116.
Quraishi UM, Pont C, Ain Qu, Flores R, Burlot L, Alaux M, et al. Combined genomic and genetic data integration of major agronomical traits in bread wheat (Triticum aestivum L). Front Plant Sci. 2017;8:1843. https://doi.org/10.3389/fpls.2017.01843.
Maccaferri M, Cane MA, Sanguineti MC, Salvi S, Colalongo MC, Massi A, et al. A consensus framework map of durum wheat (Triticum durum Desf) suitable for linkage disequilibrium analysis and genome-wide association map**. BMC Genom. 2014;15:873.
Luo MC, Gu Y, Puiu D, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2017;551:498–502. https://doi.org/10.1038/nature24486.
Marone D, Laido G, Gadaleta A, Colasuonno P, Ficco DB, Giancaspro A, et al. A high-density consensus map of A and B wheat genomes. Theor Appl Genet. 2012;125:1619–38.
Nannuru VKR, Windju SS, Belova T, et al. Genetic architecture of fusarium head blight disease resistance and associated traits in Nordic spring wheat. Theor Appl Genet. 2022;135:2247–63. https://doi.org/10.1007/s00122-022-04109-9.
Leppink J, O’Sullivan P, Winston K. Effect size-large medium and small. Perspect Med Edu. 2016;5(6):347–9. https://doi.org/10.1007/s40037-016-0308-y.
Malla S, Ibrahim AMH, Glover KD. Diallel analysis of Fusarium head blight resistance in wheat. J Crop Improv. 2009;23:213–34.
Cordell JH. Epistasis: what it means what it doesn’t mean and statistical methods to detect it in humans. Hum Mol Genet Oxford University Press. 2002;11:2463–68.
Miedaner T, Schneider B, Geiger HH. Deoxynivalenol (DON) content and Fusarium head blight resistance in segregating populations of winter rye and winter wheat. Crop Sci. 2003;43:519–26.
Geddes-McAlister J, Eudes F, Laroche A, Selinger L. Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host Hordeum vulgare. Proteom. 2008;8:545–54. https://doi.org/10.1002/pmic.200700115.
Miedaner T, Schulthess AW, Gowda M, Reif J, Longin F. High accuracy of predicting hybrid performance of Fusarium head blight resistance by mid-parent values in wheat. Theor Appl Genet. 2017. https://doi.org/10.1007/s00122-016-2826-8.
Clark AJ, Sarti-Dvorjak D, Brown-Guedira G, Dong Y, Baik BK, Van Sanford DA. Identifying rare FHB-resistant segregants in intransigent backcross and F2 winter wheat populations. Front Microbiol. 2016;7:277. https://doi.org/10.3389/fmicb.2016.00277.
Prat N, Guilbert C, Prah U, Wachter E, Steiner B, Langin T, Robert O, Buerstmayr H. QTL map** of Fusarium head blight resistance in three related durum wheat populations. Theor Appl Genet. 2017;130(1):13–27. https://doi.org/10.1007/s00122-016-2785-0.
Tessmann EW, Van Sanford DA. GWAS for fusarium head blight related traits in winter wheat (Triticum Aestivum L) in an artificially warmed treatment. Agronomy. 2018;8(5):68. https://doi.org/10.3390/agronomy8050068.
Zhao M, Leng Y, Chao S, Xu SS, Zhong S. Molecular map** of QTL for Fusarium head blight resistance introgressed into durum wheat. Theor Appl Genet. 2018;131(9):1939–51. https://doi.org/10.1007/s00122-018-3124-4.
Moreno-Amores J, Michel S, Miedaner T, et al. Genomic predictions for Fusarium head blight resistance in a diverse durum wheat panel: an effective incorporation of plant height and heading date as covariates. Euphytica. 2020;216:22. https://doi.org/10.1007/s10681-019-2551-x.
Shen XR, Ittu M, Ohm HW. Quantitative trait loci conditioning resistance to Fusarium head blight in wheat line F201R. Crop Sci. 2003;43:850–7. https://doi.org/10.2135/cropsci2003.0850.
Liu S, Abate ZA, Lu H, Musket T, Davis GL, McKendry AL. QTL associated with Fusarium head blight resistance in the soft red winter wheat Ernie. Theor Appl Genet. 2007;115:417–27. https://doi.org/10.1007/s00122-007-0577-2.
Liu J, Li L, Foroud NA, Gong X, Li C, Li T. Proteomics of bulked rachides combined with documented QTL uncovers genotype nonspecific players of the fusarium head blight responses in wheat. Phytopath. 2019;109:111–9. https://doi.org/10.1094/PHYTO-03-18-0086-R.
Gervais L, Dedryver F, Morlais JY, Bodusseau V, Negre S, Bilous M, et al. Map** of quantitative trait loci for field resistance to Fusarium head blight in an European winter wheat. Theor Appl Genet. 2003;106:961–70.
Paillard S, Schnurbusch T, Tiwari R, Messmer M, Winzeler M, et al. QTL analysis of resistance to Fusarium head blight in Swiss winter wheat (Triticum aestivum L). Theor Appl Genet. 2004;109:323–32.
Goddard R, Steed A, Chinoy C, Ferreira JR, Scheeren PL, Maciel JLN, et al. Dissecting the genetic basis of wheat blast resistance in the Brazilian wheat cultivar BR 18-Terena. BMC Plant Biol. 2020. https://doi.org/10.1186/s12870-020-02592-0.
Oettler G, Heinrich N, Miedaner T. Estimates of additive and dominance effects for Fusarium head blight resistance of winter triticale. Plant Breed. 2004;123:525–30.
Jiang Y, Yuan Z, Hu H, Ye X, Zheng Z, Wei Y, et al. Differentiating homoploid hybridization from ancestral subdivision in evaluating the origin of the D lineage in wheat. New Phytol. 2020;228:409–14.
Thambugala D, Brûlé-Babel AL, Blackwell BA, Fedak AJ, Foster D, MacEachern J, et al. Genetic analyses of native Fusarium head blight resistance in two spring wheat populations identifies QTL near the B1, Ppd-D1, Rht-1, Vrn-1, Fhb1, Fhb2, and Fhb5 loci. Theor Appl Genet. 2020;133:2775–96. https://doi.org/10.1007/s00122-020-03631-y.
Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, et al. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f sp tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L). Genes. 2015;5:4.
Kolb FL, Bai GH, Muehlbauer GJ, Anderson JA, Smith KP, Fedak G. Host plant resistance genes for Fusarium head blight: map** and manipulation with molecular markers. Crop Sci. 2001;41:611–9.
Vales MI, Schon CC, Capettini F, Chen XM, Corey AE, Mather DE, Mundt CC, Richardson KL, Sandoval-Islas JS, Utz HF, Hayes PM. Effect of population size on the estimation of QTL: a test using resistance to barley stripe rust. Theor Appl Genet. 2005;111:1260.
Schnurbusch T, Paillard S, Fossati D, Messmer M, Schachermayr G, et al. Detection of QTLs for Stagonospora glume blotch resistance in Swiss winter wheat. Theor Appl Genet. 2003;107:1226–34.
Shankar M, Walker E, Golzar H, Loughman R, Wilson R, Francki M. Quantitative trait loci for seedling and adult plant resistance to Stagonospora nodorum in wheat. Phytopath. 2008;98:886–93. https://doi.org/10.1094/PHYTO-98-8-0886.
Risser P, Ebmeyer E, Korzun V, Hartl L, Miedaner T. Quantitative trait loci for adult-plant resistance to Mycosphaerella graminicola in two winter wheat populations. Phytopath. 2011;101:1209–16.
Singh S. Map** of a resistance gene effective against Karnal bunt pathogen of wheat. Theor Appl Genet. 2003. https://doi.org/10.1007/s00122-002-1112-0.
Wang L, Cui F, Ding AM, Li J, Wang JP, Zhao CH, et al. Length of internode and spike: how do they contribute to plant height of wheat at an individual QTL level? Cere Res Commun. 2012. https://doi.org/10.1556/CRC.2012.0018.
Liu ZZ, Wang ZY. Improved scab resistance in China: Source of resistance and problems. In: Saunders DA, editor. Wheat for Nontraditional Warm Areas. CIMMYT: Texcoco, MX, USA; 1991. pp. 178–188.
Niwa S, Kubo K, Lewis J, Kikuchi R, Manickavelu A, Ban T. Variations for Fusarium head blight resistance associated with genomic diversity in different sources of the resistant wheat cultivar ‘Sumai3.’ Breed Sci. 2014;64:90–6. https://doi.org/10.1270/js.64.90.
Wang R, Liu Y, Isham KW, Zhao J, Wheeler N, Klassen Y, et al. QTL identification and KASP marker development for productive tiller and fertile spikelet numbers in two high-yielding hard white spring wheat cultivars. Mol Breed. 2018;38:135. https://doi.org/10.1007/s11032-018-0865-9.
Osman M, He X, Singh R, Duveiller E, Lillemo M, Pereyra S, et al. Phenotypic and genotypic characterization of CIMMYT’s 15th international Fusarium head blight screening nursery of wheat. Euphytica. 2015;205:279–89. https://doi.org/10.1007/s10681-015-1425-0.
Liu S, Abate ZA, McKendry AL. Inheritance of Fusarium head blight resistance in the soft red winter wheat Ernie. Theor Appl Genet. 2005;110:454–61. https://doi.org/10.1007/s00122-004-1852-0.
McKendry AL, Berg JE, Tague DN, Kephart KD. Registration of ‘Ernie’ wheat. Crop Sci. 1995;35:1513. https://doi.org/10.2135/cropsci1995.0011183X003500050061x.
Gooding RW, Lafever HN, Campbell KG, Herald LD. Registration of freedom wheat. Crop Sci. 1997;37:1007. https://doi.org/10.2135/cropsci1997.0011183X003700030057x.
Griffey CA, Starling TM, Price AM, Sisson WL, Das MK, Pridgen TH, et al. Registration of “Roane” wheat. Crop Sci. 2001;41:1359–60. https://doi.org/10.2135/cropsci2001.4141359x.
Rowland T, Perry BI, Upthegrove R, Barnes N, Chatterjee J, Gallacher D, Marwaha S. Neurotrophins, cytokines, oxidative stress mediators and mood state in bipolar disorder: systematic review and meta-analyses. Br J Psychiatry. 2018;213(3):514–27.
Ashman RB. Genetic determination of susceptibility and resistance in the pathogenesis of Candida albicans infection. FEMS Immunol Med Microbiol. 1997;19(3):183–9. https://doi.org/10.1111/j.1574-695X.1997.tb01087.x.
Stuthman DD, Leonard KJ, Miller-Garvin J. Breeding crops for durable resistance to disease. Classification of types of resistance from a breeder’s prospective. In: Stuthman DD, editor. Advances in agronomy, vol. 95. USA: Elsevier; 2007. p. 320–69.
Acknowledgements
The authors thank Ms. Nesreen ElDoliefy for the language editorial efforts and time she devoted to the manuscript. The authors are grateful to Dr. Ajay Kumar for the knowledge he provided during the QTL analysis that granted him authorship on the publication https://doi.org/10.1007/s10681-020-02610-0 and for some assistance in this study. The authors thank J. Underdahl, M. Abdallah, A. Walz, B. Schatz, J. Hegstad, and A. Peckrul for their technical assistance.
Funding
U.S. Department of Agriculture and U.S. Wheat and Barley Scab Initiative, 59-0206-9-066.
Author information
Authors and Affiliations
Contributions
Writing original draft, Data collection and Analysis by Ahmed ElFatih ElDoliefy. Research article conceptualization, Reviewing, Editing, and Validation of analyzed data by Ahmed ElFatih ElDoliefy and Mohamed Mergoum. Methodology by Ahmed ElFatih ElDoliefy, James Anderson, Karl Glover, Elias M. Elias, and Mohamed Mergoum. Software analysis by Ahmed ElFatih ElDoliefy and Ibrahim M. ElZahaby.; Formal analysis Ahmed ElFatih ElDoliefy, Elias M. Elias, Hisham A. Ashry, and Mohamed Mergoum. Data curation Ahmed ElFatih ElDoliefy, Elias M. Elias, Hisham A. Ashry, Ibrahim M. ElZahaby. Visualization by Ahmed ElFatih ElDoliefy and Ibrahim M. ElZahaby. Funding acquisition, and project administration by Mohamed Mergoum. All authors have read and agreed to the published version of the manuscript.”
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The plants used in the study is owned to the breeder who generated them who is Dr. Mohamed Mergoum (one of the authors) who is the director of the breeding program at NDSU during the time the experiments and the study were taken place. Therefore, the collection of the plants used in the study complies with local or national guidelines with no need for further affirmation. The material used in this study was based upon work supported by the U.S. Department of Agriculture, under Agreement No. 59-0206-9-066. This is a cooperative project with the U.S. Wheat and Barley Scab Initiative. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Department of Agriculture.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
ElDoliefy, A.E.A., Anderson, J.A., Glover, K.D. et al. Map** of main and hidden epistatic QTL effects in spring wheat population using medium parental FHB resistance. Discov. Plants 1, 1 (2024). https://doi.org/10.1007/s44372-024-00001-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44372-024-00001-6