Abstract
Shorea hemsleyana (King) King ex Foxw is used to treat various ailments in humans. Numerous biological activities have been reported previously. The current study sought to identify S. hemsleyana phyto-derived anti-viral compounds against the SARS-CoV-2 main protease to gain insight into the molecular interactions. In the present research, nine compounds obtained from the PubChem database are examined via molecular docking. Docking experiments were conducted using the AutoDock Vina tool. The Swiss ADME and DruLito servers were used for drug-like predictions. Our research shows that the phytoconstituents of S. hemsleyana, namely, Hemsleyanol-A and Hemsleyanoside-A, may act against SARS CoV-2 main protease with the binding affinity of − 7.6 and − 6.8 kcal/mol respectively, which were further validated by molecular dynamics (MD) simulations and end-state binding energy calculations. These phytocompounds could be used in contemporary strategies to develop effective medicines from natural sources. The identified substances are potential anti-viral agents. However, in vitro studies are necessary to assess their effectiveness against SARS-CoV-2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
An outbreak of the SARS-CoV-2 virus in 2019 caused one of the worst pandemics in history, resulting in a medical catastrophe [1]. SARS-CoV-2 has also spread to people from different countries via different routes, such as travellers, resulting in a pandemic declared by the WHO [2, 3]. The primary infection symptom of this novel coronavirus (COVID-19) is pneumonia, although additional symptoms include headache, weariness, and loss of taste.
The epidemiological history of the infection was obtained from the seafood market in Wuhan, China [4]. However, the precise origin of human transmission remains unclear. Currently, NCBI GenBank recognises over 100 genome patterns from over ten countries [5]. The variation between these series was less than 1%. SARS-CoV-2 has led to significant respiratory system infections in humans, caused by β-coronaviruses via the ACE2 receptor. Chinese experts separated SARS-CoV-2 and sequenced its genome SARS-CoV-2 on January 7, 2020 [6].
To propose an effective therapy against SARS-CoV-2, a thorough understanding of the viral structure is required, which may help identify more suitable targets [7,8,9]. Some hypothesised molecular structures for COVID-19 drug development include the spike glycoprotein, the main protease, and papain-like protease [10, 11]. Suppression of these components may result in the regression of the COVID-19 malady state.
Proteases are essential components of the SARS-CoV-2 life cycle. SARS-CoV-2 enters target cells and produces two cysteine proteases, 3-chymotrypsin-like protease or main protease and papain-like protease. These enzymes are essential for the growth and spread of this pathogen as they are involved in the maturation of viral proteins [12]. As they play an important role, these proteases could be possible targets for studying COVID-19 treatment [13]. Along with targeting the active site of these proteases, some groups have identified and designed the inhibitors against the allosteric sites of the main protease as well to inhibit the protease activity of it [14].
Bioinformatics is one of the most talked-about fields that can examine the effectiveness of drugs for various illnesses, including COVID-19, and could be beneficial [15]. This can save money, expert time, and energy if computational tests are used before applying in vitro and in vivo methods. Moreover, they do not require animals [7, 16]. Scientists can use computers to examine thousands of possible therapeutic agents and determine their effectiveness against the target protein involved in disease [17, 18]. Bioinformatics has been used to produce recombinant vaccines and identifies many natural and synthetic compounds against COVID-19 [19, 20].
Medical practitioners use antimalarial, anti-HIV, and anti-influenza medications, and their combinations, to treat COVID-19. However, these treatments do not permanently cure coronavirus infection [21]. In contrast, many phytochemicals identified in the scientific literature that exhibit potential anti-viral action might be used as alternatives to inhibit the reproduction of coronavirus [22, 23]. Natural compounds with high chemical diversity have lower production costs than biotechnological products or compounds synthesised from combinatorial chemistry and possess milder or non-existent side effects than chemical drugs [24]. Many groups have identified FDA-approved natural compounds that possess potent anti-viral activity against the main protease of SARS-CoV2 [25, 26].
Though different vaccines have been developed against the SARS-CoV-2 but their reach to under developed nations is limited and their efficacy towards the newly circulating mutants is debatable. As of 8th September 2022, World Health Organization (WHO) (https://www.who.int/activities/tracking-SARS-CoV-2-variants) and European Centre for Disease Prevention and Control (ECDC) (https://www.ecdc.europa.eu/en/covid-19/variants-concern) have assessed and kept the Omicron variants (BA.1, BA.2, BA.3, BA.4, BA.5 and XE variants) from Pango lineage B.1.1.529 as the variants of concern owing to their increased transmissibility and severity. The continuous evolution of SARS-CoV-2, especially under chronic infections in immunocompromised patients gives it sufficient time to gain advantageous mutations and makes it more resistant to current antivirals like Paxlovid (nirmatrelvir–ritonavir), molnupiravir and remdesivir and escape of recognition by the circulating antibodies [27,28,29]. Due to the development of immune-evasion and incidences of resistance to current antivirals against SARS-CoV-2, there is a dire need to continuously develop effective vaccines and identify new antivirals against the SARS-CoV-2. To fill this gap of identifying effective antivirals, our study is designed to screen novel phytocompounds, through a structure-based drug discovery approach employing molecular docking-based virtual screening against the main protease (Mpro) of SARS-CoV-2, followed by validation of docking results using MD simulations. The findings of this study can be used to select potential therapeutic candidates for in-vitro, in-vivo, and clinical testing.
2 Materials and Methods
2.1 Preparation of Ligands
A list of active phytochemicals was acquired through literature review [30, 31]. Nine active compounds from Shorea hemsleyana, i.e., Hemsleyanol-B (Pubchem ID: 10842394), Hemsleyanoside-D (Pubchem ID: 101073881), Hopeaphenol (Pubchem ID: 495605), Hemsleyanoside-A (Pubchem ID: 101073244), Davidiol-A (Pubchem ID: 11614520), Hemsleyanoside-B (Pubchem ID: 101073245), Hemsleyanoside-C (Pubchem ID: 101073880), Hemsleyanol-A (Pubchem ID: 10814213), and Resveratrol-12-C-beta-glucopyranoside (Pubchem ID: 101011049) were retrieved from the PubChem database as shown in Table 1. This database was used to obtain the three-dimensional structures of these bioactive chemicals in SDF format. In addition, the SDF structures were translated into PDB format using PyMol software. Ligands were protonated suitable to pH 7.4, and energy minimised using a conjugate gradient algorithm for 500 steps with a step size of 0.02 Å updated after every 10 steps.
2.2 Preparation of Protein
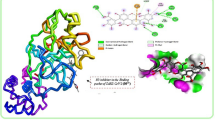
The X-ray crystal structure of the SARS CoV-2 main protease (PDB ID 6LU7) was retrieved from the RCSB Protein Data Bank (Fig. 1). MGL AutoDock Tools were used to prepare the protein, which included the removal of crystal water and ligands and the addition of Kollman charges and polar hydrogens [34].
2.3 Drug-likeliness and ADMET Analysis
The phytochemical compounds were obtained from PubChem in SDF format and subjected to drug-likeness predictions using DruLiTo software [35, 36]. The pharmacokinetic properties of ligands must be investigated to establish their roles in the body. The ADMET profiles of the ligands were studied using the Swiss ADME, admetSAR, and ProTox-II web servers [37, 38].
2.4 Active Site Prediction
A critical step is to predict the active sites of a target by using computational tools. Supercomputing Facility for Bioinformatics and Computational Biology, IIT Delhi (scfbio-iitd.res.in) was used to extract information about the active site of the main protease structure file used (PDB ID:6LU7), and it was visualised in BIOVIA Discovery Studio Visualizer 2020 (Fig. 2).
2.5 Compound Screening Using the PyRx Program
After loading macromolecules and ligands in the PyRx program (0.9.8), they were converted and saved in the pdbqt format. The grid parameter configuration file was generated using PyRx, with the dimensions of the grid box extracted from co-crystal inhibitor N3 complexed with 6LU7. Autodock Vina was chosen as the docking algorithm with exhaustiveness set to 100. PyRx employing the AutoDock Vina algorithm is fast and precise in stable conformer identification. It is offered in normal, reverse, or combined docking mode. Molecular docking was performed according to the Wizard’s stage [39]. The stable docked pose conformations of the ligands were selected based on docking binding energy scores and non-bonded interactions with critical active site residues. The docking protocol was also validated by redocking the co-crystal ligand N3 onto the main protease 6LU7 and overlapped with the native crystal pose.
2.6 Analysis and Visualisation
The Vina score data presented in kcal/mol were compared between the target, co-crystal inhibitor, and phytocompounds. The three-dimensional conformations of the docked ligands in pdbqt format were merged, analysed, and visualised using BIOVIA Discovery Studio Visualizer 2020 [40, 41].
2.7 Molecular Dynamics (MD) Simulations
The screened molecules obtained after the docking simulations were subjected to all-atom MD simulations using LiGRO [42], an automated GUI-based tool to prepare a system for running simulations using GROMACS 5.1.5 [43] version. The separate protein-ligand complex systems were solvated in a cubic box with dimensions of 893.27 nm^3 solvated with explicit transferable intermolecular potential with 3 points (TIP3P) water molecules. The topology of the ligand was generated using ACPYPE with a general Amber force field (GAFF), and the BCC model was used for charge calculations. The AMBER99SB force field was used for protein topology. The protein has 306 residues and a charge of – 4, and the system was neutralised using 0.15 M NaCl concentration, adding 85 Na+ and 81 Cl− ions. The prepared system was energy-minimised to remove any steric clashes under 1000 steepest descent steps, followed by 200 conjugate gradient steps (Supplementary Fig. 1a and 2a show convergence of Potential energy of system to minimum over minimisation steps). The minimised system was equilibrated under NVT (modified Berendson Thermostat) and NPT (Parinello-Rahman Barostat) ensembles for 1ns each (Supplementary Figs. 1b–d and 2b–d show equilibrium of the system evident from the system Temperature, Pressure and water density plots over the NVT and NPT runs), followed by a production run of 100 ns under the NPT ensemble. A neighbor search was performed using the Verlet cutoff scheme, and cutoff values of 1.4 nm were used for short-range electrostatic and van der Waals energies. Long-range electrostatics were managed using the PME method, and the LINCS algorithm constrained covalent bonds. A time step integrator of 2 fs was used, and 10,000 frames were saved over the 100 ns simulation time. The trajectory was visualised using VMD and analyzed using standard GROMACS tools such as RMSD, RMSF, Rg, and H-bonds. The interaction fraction of the ligand with the active site residues of 6LU7 was calculated using the Molecular-dynamics-Interaction-plot tool (https://github.com/tavolivos/Molecular-dynamics-Interaction-plot) for 500 equidistant frames extracted from the trajectory.
The end-state MMGBSA binding free energy was calculated for the protein-ligand complexes using the gmx_MMPBSA tool [44]. One hundred frames from each complex trajectory’s last 10 ns of the metastable region were selected for binding free energy calculations. iGB model 2 and the internal dielectric constant value set to 1. The binding free energy was calculated using the following Eq. 1:
∆G gas refers to the total gas-phase energy consisting of electrostatic and van der Waals interaction energies, Gsolv refers to the polar and non-polar solvation free energies, and TΔS is the change in interaction entropy on ligand binding.
The metadynamics analysis of the complex MD trajectories was done using geo_measures v_0.9 [45] pymol plugin. The analyses included the Free Energy Landscape (FEL), Principal Component Analysis (PCA), and the porcupine plot. The FEL was generated as a function of the trajectory RMSD vs. RG, and the first two principal components (PC1 and PC2) were used for the PCA analysis.
3 Results and Discussion
3.1 Drug Likeliness Properties
Drug likeness is a critical screening measure for drug candidates during the drug discovery and development phases. This metric correlates the physicochemical properties of a substance with its biopharmaceutical properties in the human body, particularly its influence on oral bioavailability [46].
The DruLito program was used to study the physicochemical characteristics of the nine selected active chemicals. Except for resveratrol 12-C-beta-glucopyranoside and hemsleyanol A, the other substances did not follow Lipinski’s rule (Table 2) [47]. However, many natural compounds do not follow Lipinski’s rule of five but remain largely bioavailable and mimic the structure of biological synthetic intermediates and endogenous metabolites involved in cellular pathways [48].
3.2 ADMET Evaluation of the Phytocompounds
The ADMET attributes of the ligands have been studied using Swiss ADME (http://www.swissadme.ch/), admetSAR (http://lmmd.ecust.edu.cn/admetsar2/) and Protox-II (https://tox-new.charite.de/protox_II/) web servers. Table 3 represents the predicted results of the ADMET properties of the selected phytocompounds.
Ideal drug candidates should be non-toxic and exhibit acceptable ADME characteristics. Using SwissADME, the ADME profile of the identified molecules, including drug-likeness, partition coefficient, solubility, HIA, BBB, and cytochrome P450 inhibition, was examined. At the early stages of drug discovery and development, drug likeness is a vital criterion to be considered. It involves correlating the physicochemical properties of a compound to its biopharmaceutical properties, particularly its influence over bioavailability via oral administration [46].
The ability to absorb drugs through the human gut [HIA] is one of the most important properties of ADMET. HIA plays an important role in the transport of drugs to their target [49]. When HIA is higher, the compound will be better absorbed by the intestinal tract. Aside from Hemsleyanoside B, all compounds showed greater HIA values than 0.9, indicating good membrane permeation. The penetration power of Hemsleyanoside A and Resveratrol 12-C-beta-glucopyranoside was low compared to other phytoconstituents across the Blood Brain Barrier (BBB). In terms of predicting the efflux by P-glycoprotein from the cell, Hemsleyanoside B, Hemsleyanoside C, Hemsleyanoside D, and Resveratrol 12-C-beta-glucopyranoside come out to be non-inhibitor and substrate of P-glycoprotein, whereas, Davidiol A, Hemsleyanol A, Hemsleyanol B, and Hopeaphenol comes out to be non-inhibitor and non-substrate. An inhibitor of P-glycoprotein means that the drug will inhibit the cell’s efflux process and enhance its bioavailability. A non-inhibitor of P-glycoprotein means that the drug will efflux from the cell by P-glycoprotein and limits the bioavailability by pum** back into the lumen and may promote the elimination of that drug into the bile and urine. In the case of metabolism, Davidiol A, Hemsleyanol A, Hemsleyanol B, and Hopeaphenol were found to be non-inhibitor and substrates, while Hemsleyanoside A, Hemsleyanoside B, Hemsleyanoside C, Hemsleyanoside D, and Resveratrol 12-C-beta-glucopyranoside found to be non-inhibitor and non-substrate. A non-inhibitor of cytochrome P450 means that the molecule will not hinder the biotransformation of the compound (drug) metabolised by cytochrome P450.
Lipinski and Veber [50, 51] rule-based filters were used to calculate the drug- and lead likeness for the nine selected compounds. The results showed that all selected compounds had violated the underlying drug-likeness rules, except Hemsleyanol A and Resveratrol 12-C-beta-glucopyranoside. These results suggest that these compounds have low theoretical oral bioavailability, according to Lipinski’s rule of five. Another important property of oral drugs is their solubility in intestinal fluid because insufficient solubility can limit intestinal absorption through the portal vein system. All the compounds with low aqueous solubility levels are shown in Table 3, except for compounds Hemsleyanoside A, Hemsleyanoside B, and Resveratrol 12-C-beta-glucopyranoside, which have shown better solubility. Most of the compounds found are unlikely to cross the BBB. No compound shows a good likelihood of being a BBB penetrant.
Pan-Assay Interference Compounds (PAINS) are well-known to medicinal chemists who have spent many hours optimising these nonprogressive compounds without success. Table 3 demonstrates that none of the tested substances generated a PAINS warning.
The cytochrome P450 family facilitates drug elimination through metabolic biotransformation. Inhibiting these isoenzymes is undoubtedly a high-risk cause of pharmacokinetic interactions, leading to toxic or unwanted side effects due to a decreased clearance and accumulation of the drug or its metabolites. As shown in Table 3, all the compounds are non-inhibitors of CYP1A2, CYP2C19, CYP2C9, and CYP3A4 and, therefore, may have no side effects (such as liver dysfunction). The CYP1A2 enzyme is found predominantly in the liver (about 10% of the total CYP content) and is responsible for activating amines, PAHs, and many other drugs [52].
These compounds have been evaluated for their hepatotoxicity, carcinogenicity, mutational potential, and cytotoxicity [53]. According to the results of ProTox II, The Carcinogenic profile also shows that except for Hemsleyanol B and Hopeaphenol, remaining all the compounds were non-carcinogenic, so they can be applied as drugs for treating COVID-19 as there would not be any bioaccumulation of compounds in the human body and these compounds less likely to cause cancer in future if the patient were treated for a long duration. Immunotoxicity has been reported for Hemsleyanol B, Hemsleyanoside A, Hemsleyanoside B, Hemsleyanoside C, and Resveratrol 12-C-beta-glucopyranoside. No hepatotoxicity or cytotoxicity is associated with any of the compounds (Table 3).
The LD50 prediction using ProTox II indicated that compounds, except Hemsleyanoside A, have non-toxic effects in rats and have oral LD50 values ranging from 250 to 10,000 mg/kg.
3.3 Molecular Docking Studies
The docking procedures were validated via re-docking the co-crystallised ligand (N3) against the active pocket of the binding site. The calculated RMSD values between the re-docked pose and the co-crystallised one were 0.375Aº, respectively. Such values of RMSD indicated the efficiency and validity of the docking protocol (Fig. 3). The binding site was discovered to be predominantly localised in the hydrophobic gap bordered by the following amino acids using the inhibitor N3 with 6lu7: PHE A:140, GLY A:143, HIS A:164, GLN A:189, LEU A:167, MET A:165, THR A:190, ALA A:191, PRO A:168, GLU A:166, MET A:49 and HIS A:41.
There were eight hydrogen bond interactions with eight amino acids, two with GLU A:166 and six with PHE A:140, GLY A:143, HIS A:163, HIS A:164, GLN A:189, and THR A:190, as shown in Fig. 1. Two C-H bonds exist with MET A:165 and HIS A:172, as well as three hydrophobic Pi-alky bonds with PRO A:168, ALA A:191, and HIS A:41. Subsequent investigation revealed the existence of an Amide-Pi stacking interaction with LEU A:141 (Fig. 4).
The redocked pose of the co-crystallised N3 and the co-crystallised ligand interacting with similar amino acid residues of the active site, as shown in Fig. 4.
In order to identify potential inhibitor candidates against the main protease of SARS CoV-2, molecular docking of nine phytoconstituents obtained from Shorea hemsleyana was performed on the main protease and evaluated the molecular interactions based on molecular docking findings.
A structural conformation study of the ligand-protein complexes was performed to identify the target’s drug surface hotspot, and ligand-bounded amino acid residues were also identified. The compounds having the highest molecular docking scores with the Mpro SARS-CoV-2 protein are included in Table 4.
Based on the molecular docking investigation results, only three molecules demonstrated the highest binding energy values close to the co-crystallised ligand. These findings indicated that the three investigated compounds might act as potential SARS-CoV-2 inhibitors.
As depicted in Fig. 5a, Hemsleyanol-A has showed the following interaction types with the Mpro protein of SARS-CoV-2 with a binding energy of – 7.6 Kcal/mol: three H-bonds GLU A:166 (4.25), PHE A:140 (4.54), HIS A:172 (5.40), two hydrophobic interactions with LEU A:141 (7.20), CYS A:145 (7.08) and one electrostatic interaction with MET A:49 (5.62) residues.
Although Hemsleyanol-B binds to the main protease with an energy value of – 6.8 kcal/mol; herein, it shows five H-bond interactions which maintain the stability of the complex, i.e., HIS A:41 (5.07), HIS A:164 (5.72), ASN A:142 (4.61), SER A:46 (2.48), GLU A:166 (3.86, 4.66); one hydrophobic interaction with LEU A:141 (7.67) and one electrostatic interaction with CYS A:145 (7.18) (Fig. 5b).
Resveratrol-12-C-beta-glucopyranoside, isolated from S. hemsleyana, exhibits a binding energy of − 6.1 kcal/mol with the SARS CoV-2 Mpro protein. This compound interacts with the Mpro protein through H-bond with HIS A:163 (5.21), CYS A:145 (3.94), SER A:144 (2.82, 3.80), LEU A:141 (5.78), GLY A:143 (3.45), HIS A:164 (6.49) amino acid residue and hydrophobic interactions with GLN A:189 (3.58). The interaction modes are illustrated in Fig. 5c.
These amino acid residues in the Mpro protein, GLU A:166, and LEU A:141, can contribute substantially to Hemsleyanol-A and Hemsleyanol-B stability.
3.4 MD Simulations
The dynamic native-like behavior of the protein-ligand complex was mimicked using all-atom MD simulations ran for 100 ns. The effect of solvation on the interaction between the target protein and the screened ligands was assessed, which was not considered during the docking protocol. Different studies have validated the time duration required for assessing the stable interactions between protein-ligand by running MD simulation for 100 ns and compared with µs simulation results where no significant difference was observed in prolonged simulations for the stable binding ligand compounds. In contrast, for non-stable complexes, longer simulation is recommended [54, 55]. The stability of the 6LU7 main protease protein with the screened molecules (Hemsleyanoside-A and Hemsleyanol-A) was assessed by monitoring the backbone Root Mean Square Deviation (RMSD) of the free and ligand-bound protein. As evident from Fig. 6a, both the systems were well equilibrated and stable, with the average RMSD for the 6LU7-Hemsleyanoside-A complex being 2.65 Å and the 6LU7-Hemsleyanol-A complex was also stable, with the average RMSD of 2.4 Å up to 80 ns. It increased up to 3.7 Å due to the increased movement of the end C-terminal as evident from the Root Mean Square Fluctuation (RMSF) plot (Fig. 6b). The overall fluctuation in the active site residues for both the complexes was well within 2.4 Å. The Radius of Gyration (RG) of the protein complexes was stable (Fig. 6c), with contraction of the protein being observed during the last 20 ns of the 6LU7-Hemsleyanol-A complex. Both the ligands formed 6–7 hydrogen bonds (Fig. 6d) with the active site residues of the protein. The highest occupancy of the hydrogen bonds with the cutoff distance of 3.5 Å was observed with residues THR190 (52.55%), GLU166 (22.85%), GLY143 (21.27%), ARG188 (7.14%), and SER144 (5.81) along with other residues having lower occupancies in case of 6LU7- Hemsleyanoside-A complex. In contrast, in the case of the 6LU7-Hemsleyanol-A complex, the highest occupancy for the hydrogen bonds was observed with the residues ASP187 (67.77%), GLU166 (91.69%), HIS164 (13.89%), GLN192 (7.57%), along with other residues with occupancy up to 1% were considered (Table 5).
The distance and contact analysis of the residues within 3.5–5 Å of the ligand molecules was also done. From Figs. 7 and 8 show that both the ligands formed contacts with up to 5 active site residues, which were within 3.5 Å distance from the ligand molecules, with them being under 3.5 Å from 90 to 98% of the trajectory time. Figures 7d and 8d show the significant interactions of the residues lining the active site (within 5 Å of the ligand) and their interaction types. In both cases, major interactions were hydrogen bonds, followed by hydrophobic and pi-stacking interactions. Overall, the analysis of these results suggested that the complexes of main-protease with Hemsleyanoside-A and Hemsleyanol-A attained stable equilibrium under the time frame of 100 ns of the study.
Distance-based interaction analysis of active site residues of main protease within 3.5 Å of Hemsleyanoside-A. a Residues maintain a 3.5 Å distance with the ligand over 100 ns simulation trajectory, b total number of contacts within 3.5 Å of the ligand, c total number of contacts formed between the active site residues within 3.5 Å of the ligand over the 10,000 frames of the MD trajectory, d interaction fraction along with the type of non-bonding interactions between the active site residues and the ligand. Note: LIG 307 here refers to Hemsleyanoside-A
Distance-based interaction analysis of active site residues of main protease within 3.5 Å of Hemsleyanol-A. a Residues maintain a 3.5 Å distance with the ligand over 100 ns simulation trajectory, b total number of contacts within 3.5 Å of the ligand, c total number of contacts formed between the active site residues within 3.5 Å of the ligand over the 10,000 frames of the MD trajectory, d interaction fraction along with the type of non-bonding interactions between the active site residues and the ligand. Note: LIG 307 here refers to Hemsleyanol-A
The metadynamics analysis of the complex trajectories was done to map the different conformational ensemble states spanned by the protein in the complex with the screened ligands. The FEL (Fig. 9a) of the Hemsleyanoside-A complex showed a single global energy minimum at RMSD of 0.18 nm and RG 2.2 nm, while the 6LU7-Hemsleyanol-A complex, one local minimum and one diffused global energy minimum was observed with energy difference less than 2 kcal/mol. The 6LU7-Hemsleyanol-A complex spanned a comparatively larger search space in both directions along with the first principal than the Hemsleyanoside-A complex owing to the different conformation states obtained by the movement of the C-terminal stretch (Fig. 9b), as can be seen in the porcupine plot also (Fig. 9d). The first 10 PCs explained 76% and 82% of the possible movements for the Hemsleyanoside-A complex and 6LU7-Hemsleyanol-A complex (Fig. 9c).
The binding free energy analyses (Figs. 10 and 11) showed that the relative binding free energy for both the ligands with the target protein was similar (– 52.83 ± 3.52 kcal/mol for Hemsleyanoside-A complex and − 52.78 ± 3.85 kcal/mol for 6LU7-Hemsleyanol-A complex). Both the ligands remained stably bound throughout the trajectory, and the major contributing active site residues in the binding energy are shown in Figs. 10e and 11e.
End-state MMGBSA binding energy analysis between main protease and Hemsleyanoside-A. a Enthalpic contributions to the binding free energy, b entropic contributions to the binding free energy, c total binding free energy, d total decomposition of the binding energy per selected frames of the trajectory, e per-residue decomposition of the binding energy contributions, f per-residue per-frame binding energy contributions
End-state MMGBSA binding energy analysis between main protease and Hemsleyanol-A. a Enthalpic contributions to the binding free energy, b entropic contributions to the binding free energy, c total binding free energy, d total decomposition of the binding energy per selected frames of the trajectory, e per-residue decomposition of the binding energy contributions, f per-residue per-frame binding energy contributions
4 Conclusion
Despite massive attempts to curb the current epidemic, the coronavirus disease (COVID-19) pandemic is spreading worldwide. Although many people worldwide are unvaccinated, several vaccines are being researched and authorised for emergency use, particularly among individuals in underdeveloped countries. The development of anti-viral medications may help to combat this pandemic. We conducted an in silico investigation to identify potential inhibitors of SARS-CoV-2 main protease. The research used Nine molecules obtained from S. hemsleyana, a Dipterocarpaceae species. Only seven of the nine compounds had good binding affinities in the molecular docking investigation compared to the reference co-crystallised ligand N3. The overall binding energies of the nine compounds ranged between − 3.1 and − 7.6 kcal/mol, which is close to the value found for the co-crystallised ligand (-8.2 kcal/mol). ADMET analysis was performed on nine selected drugs to determine their absorption, distribution, metabolism, and toxicity. Molecular dynamics (MD) simulations were performed to confirm the stability of hemsleyanol-A and hemsleyanoside-A against the main protease. The acquired findings showed that both compounds interacted with the active site residues, maintaining a distance of less than 3.5 Å over the simulated time. Based on the findings of this study and earlier research, hemsleyanol-A and hemsleyanoside-A might be explored as potential inhibitors of the SARS-CoV-2 main protease. Experiments should be conducted to determine in vitro and in vivo efficacy against SARS-CoV-2.
References
Ram H, Sharma R, Dewan D, Singh A (2020) Insight view on pandemic COVID-19: worldwide perspective with homoeopathic management through repertorial analysis. Homœopath Links 33(03):180–195
Malik YS, Kumar N, Sircar S, Kaushik R, Bhat S, Dhama K et al (2020) Coronavirus Disease Pandemic (COVID-19): challenges and a global perspective. Pathogens 9(7):519. https://doi.org/10.3390/pathogens9070519
Covid C, Team R, Covid C, Team R, Bialek S, Gierke R et al (2020) Coronavirus disease 2019 in children—United States, february 12–april 2, 2020. Morb Mortal Wkly Rep 69(14):422
Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y et al (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395(10223):507–513. https://doi.org/10.1016/S0140-6736(20)30211-7
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y et al (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med 382(13):1199–1207. https://doi.org/10.1056/NEJMoa2001316
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224):565–574. https://doi.org/10.1016/S0140-6736(20)30251-8
Mohabatkar H, Behbahani M, Moradi M (2021) A concise IN silico prediction report OF a potential PRION-like domain IN SARS-COV-2 polyprotein. J Microbiol Biotechnol Food Sci 11(3):e4813–e4813
Mohabatkar H, Ebrahimi S, Moradi M (2021) Using Chou’s five-steps rule to classify and predict glutathione S-Transferases with different machine learning algorithms and pseudo amino acid composition. Int J Pept Res Ther 27(1):309–316
Valle C, Martin B, Touret F, Shannon A, Canard B, Guillemot JC et al (2020) Drugs against SARS-CoV‐2: what do we know about their mode of action? Rev Med Virol 30(6):1–10
Wong F-C, Ong J-H, Chai T-T (2021) SARS-CoV-2 spike protein-, main protease-and papain-like-protease-targeting peptides from seed proteins following gastrointestinal digestion: an in silico study. Phytomed Plus 1(1):100016
Wong FC, Ong JH, Kumar DT, Chai TT (2021) In silico identification of multi-target anti-SARS-CoV-2 peptides from Quinoa seed proteins. Int J Pept Res Ther 27(3):1837–1847. https://doi.org/10.1007/s10989-021-10214-y
Gao X, Qin B, Chen P, Zhu K, Hou P, Wojdyla JA et al (2021) Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm Sin B 11(1):237–245. https://doi.org/10.1016/j.apsb.2020.08.014
Báez-Santos YM, John SES, Mesecar AD (2015) The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir Res 115:21–38. https://doi.org/10.1016/j.antiviral.2014.12.015
Mensah JO, Ampomah GB, Gasu EN, Adomako AK, Menkah ES, Borquaye LS (2022) Allosteric modulation of the main protease (MPro) of SARS-CoV-2 by casticin—insights from molecular dynamics simulations. Chem Afr 5:1305–1320. https://doi.org/10.1007/s42250-022-00411-7
Nabati F, Moradi M, Mohabatkar H (2020) In silico analyzing the molecular interactions of plant-derived inhibitors against E6AP, p53, and c-Myc binding sites of HPV type 16 E6 oncoprotein. Mol Biol Res Commun 9(2):71–82. https://doi.org/10.22099/mbrc.2020.36522.1483
Haghighi O (2021) In silico study of the structure and ligand preference of pyruvate kinases from Cyanobacterium Synechocystis sp. PCC 6803. Appl Biochem Biotechnol 193(11):3651–3671. https://doi.org/10.1007/s12010-021-03630-9
Ahmadi K, Farasat A, Rostamian M, Johari B, Madanchi H (2022) Enfuvirtide, an HIV-1 fusion inhibitor peptide, can act as a potent SARS-CoV-2 fusion inhibitor: an in silico drug repurposing study. J Biomol Struct Dyn 40(12):5566–5576
Gharbavi M, Johari B, Rismani E, Mousazadeh N, Taromchi AH, Sharafi A (2020) NANOG decoy oligodeoxynucleotide–encapsulated niosomes nanocarriers: a promising approach to suppress the metastatic properties of U87 human glioblastoma multiforme cells. ACS Chem Neurosci 11(24):4499–4515
Behmard E, Soleymani B, Najafi A, Barzegari E (2020) Immunoinformatic design of a COVID-19 subunit vaccine using entire structural immunogenic epitopes of SARS-CoV-2. Sci Rep 10(1):1–12
Fallah MS, Bayati M, Najafi A, Behmard E, Davarpanah S (2021) Molecular docking investigation of antiviral herbal compounds as potential inhibitors of SARS-CoV-2 spike receptor. Biointerface Res Appl Chem 11(5):12916–12924
Joshi RS, Jagdale SS, Bansode SB, Shankar SS, Tellis MB, Pandya VK et al (2021) Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn 39(9):3099–3114. https://doi.org/10.1080/07391102.2020.1760137
Elfiky AA (2021) Natural products may interfere with SARS-CoV-2 attachment to the host cell. J Biomol Struct Dyn 39(9):3194–3203
Farooq S, Ngaini Z (2021) Natural and synthetic drugs as potential treatment for coronavirus disease 2019 (COVID-2019). Chem Afr 4(1):1–13
Kitazato K, Wang Y, Kobayashi N (2007) Viral infectious disease and natural products with antiviral activity. Drug Discov Ther 1(1):14–22
Singh G, Mishra R, Goel G, Chaudhuri T (2020) Old arsenal to combat new enemy: repurposing of commercially available FDA approved drugs against main protease of SARS-CoV2. https://doi.org/10.26434/chemrxiv.13032578.v1
Prasanth D, Murahari M, Chandramohan V, Panda SP, Atmakuri LR, Guntupalli C (2021) In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J Biomol Struct Dyn 39(13):4618–4632. https://doi.org/10.1080/07391102.2020.1779129
Okpeku M (2022) Possibility of COVID-19 eradication with evolution of a new omicron variant. Infect Dis Poverty 11(1):1–3
Gandhi S, Klein J, Robertson AJ, Peña-Hernández MA, Lin MJ, Roychoudhury P et al (2022) De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat Commun 13(1):1–8
Callaway E (2022) How months-long COVID infections could seed dangerous new variants. Nature 606(7914):452–455
Chaudhuri S, Symons JA, Deval J (2018) Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antivir Res, pp 15576–15588
Mohanraj K, Karthikeyan BS, Vivek-Ananth RP, Chand RPB, Aparna SR, Mangalapandi P et al (2018) IMPPAT: a curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci Rep 8(1):4329. https://doi.org/10.1038/s41598-018-22631-z
Ito T, Tanaka T, Ido Y, Nakaya KI, Iinuma M, Riswan S (2000) Stilbenoids isolated from stem bark of Shorea hemsleyana. Chem Pharm Bull (Tokyo) 48(7):1001–1005. https://doi.org/10.1248/cpb.48.1001
Ito T, Tanaka T, Ido Y, Nakaya K, Iinuma M, Riswan S (2000) Four new stilbenoid C-glucosides isolated from the stem bark of Shorea hemsleyana. Chem Pharm Bull (Tokyo) 48(12):1959–1963. https://doi.org/10.1248/cpb.48.1959
Benhander GM, Abdusalam AAA (2022) Identification of potential inhibitors of SARS-CoV-2 main protease from Allium roseum L. molecular docking study. Chem Afr 5(1):57–67. https://doi.org/10.1007/s42250-021-00296-y
Bokka CS, Veeramachaneni GK, Thunuguntla V, Bobbillapati J, Bondili JS (2019) Peptide map**, in silico and in vivo analysis of allergenic sorghum profilin peptides. Medicina (Kaunas) 55(5):178. https://doi.org/10.3390/medicina55050178
Veeramachaneni GK, Thunuguntla V, Bhaswant M, Mathai ML, Bondili JS (2019) Pharmacophore directed screening of agonistic natural molecules showing affinity to 5HT2C receptor. Biomolecules 9(10):556. https://doi.org/10.3390/biom9100556
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G et al (2012) admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52(11):3099–3105. https://doi.org/10.1021/ci300367a
Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z et al (2019) admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 35(6):1067–1069. https://doi.org/10.1093/bioinformatics/bty707
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. In: Chemical biology. Springer, pp 243–250. Accessed 11 May 2022
Eda SR, **ka R (2019) Combined e-pharmacophore based screening and docking of PI3 kinase with potential inhibitors from a database of natural compounds. Bioinformation 15(10):709–715. https://doi.org/10.6026/97320630015709
Seeliger D, de Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24(5):417–422. https://doi.org/10.1007/s10822-010-9352-6
Kagami LP, das Neves GM, da Silva AWS, Caceres RA, Kawano DF, Eifler-Lima VL (2017) LiGRO: a graphical user interface for protein–ligand molecular dynamics. J Mol Model 23(11):1–6
Abraham M, Van Der Spoel D, Lindahl E, Hess B (2015) the GROMACS development team (2014) GROMACS User Manual version 5.0. 4.
Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E (2021) gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput 17(10):6281–6291
Kagami LP, das Neves GM, Timmers L, Caceres RA, Eifler-Lima VL (2020) Geo-Measures: a PyMOL plugin for protein structure ensembles analysis. Comput Biol Chem. https://doi.org/10.1016/j.compbiolchem.2020.107322
Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL (2012) Quantifying the chemical beauty of drugs. Nat Chem 4(2):90–98. https://doi.org/10.1038/nchem.1243
Ertl P, Rohde B, Selzer P (2000) Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem 43(20):3714–3717
Ganesan A (2008) The impact of natural products upon modern drug discovery. Curr Opin Chem Biol 12(3):306–317
Ejeh S, Uzairu A, Shallangwa GA, Abechi SE (2021) In silico design, drug-likeness and ADMET properties estimation of some substituted thienopyrimidines as HCV NS3/4A protease inhibitors. Chem Afr 4(3):563–574
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623
Di L (2014) The role of drug metabolizing enzymes in clearance. Expert Opin Drug Metab Toxicol 10(3):379–393. https://doi.org/10.1517/17425255.2014.876006
Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL et al (2012) Large-scale prediction and testing of drug activity on side-effect targets. Nature 486(7403):361–367. https://doi.org/10.1038/nature11159
De Vita S, Chini MG, Bifulco G, Lauro G (2021) Insights into the ligand binding to bromodomain-containing protein 9 (BRD9): a guide to the selection of potential binders by computational methods. Molecules 26(23):7192
Zhang Y (2020) Simulation of protein dynamics for mechanistic insight and drug design. The University of Manchester, Manchester
Acknowledgements
The authors thank the management of KVSR Siddhartha College of Pharmaceutical Sciences, Vijayawada, Krishna, Andhra Pradesh, and the HPC facility of the Indian Institute of Technology, Delhi, for providing the facilities to perform this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Prasanth, D.S.N.B.K., Singh, G., Panda, S.P. et al. In Silico Screening of Plant-Derived Anti-virals from Shorea hemsleyana (King) King ex Foxw Against SARS CoV-2 Main Protease. Chemistry Africa 6, 345–366 (2023). https://doi.org/10.1007/s42250-022-00521-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-022-00521-2