Abstract
Purpose
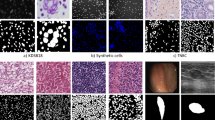
Segmentation of nuclei and cytoplasm in cellular images is essential for estimating the prognosis of lung cancer disease. The detection of these organelles in the unstained brightfield microscopic images is challenging due to poor contrast and lack of separation of structures with irregular morphology. This work aims to carry out semantic segmentation of nuclei and cytoplasm in lung cancer brightfield images using the Swin-Unet Transformer.
Methods
For this study, publicly available brightfield images of lung cancer cells are pre-processed and fed to the Swin-Unet for semantic segmentation. Model specific hyperparameters are identified after detailed analysis and the segmentation performance is validated using standard evaluation metrics.
Results
The hyperparameter analysis provides the selection of optimum parameters as focal loss, learning rate of 0.0001, Adam optimizer, and Swin Transformer patch size of 4. The obtained results show that with these parameters, the Swin-Unet Transformer accurately segmented the nuclei and cytoplasm in the brightfield images with pixel-F1 scores of 90.71% and 79.29% respectively.
Conclusion
It is observed that the model could identify nuclei and cytoplasm with varied morphologies. The detection of cytoplasm with weak and subtle edge details indicates the effectiveness of shifted window based self attention mechanism of Swin-Unet in capturing the global and long distance pixel interactions in the brightfield images. Thus, the adopted methodology in this study can be employed for the precise segmentation of nuclei and cytoplasm for assessing the malignancy of lung cancer disease.








Similar content being viewed by others
Data Availability
The data used in the manuscript is from a publicly accessible dataset and the details are provided in Sect. 2.1.
References
Pramanik, S. K., & Das, A. (2021). Fluorescent probes for imaging bioactive species in subcellular organelles. Chemical Communications, 57(91), 12058–12073. https://doi.org/10.1039/D1CC04273D
Balachandra, S., Sarkar, S., & Amodeo, A. A. (2022). The nuclear-to-cytoplasmic ratio: Coupling DNA content to cell size, cell cycle, and biosynthetic capacity. Annual Review of Genetics, 56, 165–185. https://doi.org/10.1146/annurev-genet-080320-030537
Svenningsen, E. B., & Poulsen, T. B. (2019). Establishing cell painting in a smaller chemical biology lab–A report from the frontier. Bioorganic & Medicinal Chemistry, 27(12), 2609–2615. https://doi.org/10.1016/j.bmc.2019.03.052
Kobayashi, H., Lei, C., Wu, Y., Mao, A., Jiang, Y., Guo, B., & Goda, K. (2017). Label-free detection of cellular drug responses by high-throughput bright-field imaging and machine learning. Scientific Reports, 7(1), 12454. https://doi.org/10.1038/s41598-017-12378-4
Wang, R., Butt, D., Cross, S., Verkade, P., & Achim, A. (2023). Bright-field to fluorescence microscopy image translation for cell nuclei health quantification. Biological Imaging, 3, e12. https://doi.org/10.1017/S2633903X23000120
Cross-Zamirski, J. O., Mouchet, E., Williams, G., Schönlieb, C. B., Turkki, R., & Wang, Y. (2022). Label-free prediction of cell painting from brightfield images. Scientific Reports, 12(1), 10001. https://doi.org/10.1038/s41598-022-12914-x
Fishman, D., Salumaa, S. O., Majoral, D., Laasfeld, T., Peel, S., Wildenhain, J., & Parts, L. (2021). Practical segmentation of nuclei in brightfield cell images with neural networks trained on fluorescently labelled samples. Journal of Microscopy, 284(1), 12–24. https://doi.org/10.1111/jmi.13038
Ounkomol, C., Seshamani, S., Maleckar, M. M., Collman, F., & Johnson, G. R. (2018). Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nature Methods, 15(11), 917–920. https://doi.org/10.1038/s41592-018-0111-2
Hollandi, R., Moshkov, N., Paavolainen, L., Tasnadi, E., Piccinini, F., & Horvath, P. (2022). Nucleus segmentation: Towards automated solutions. Trends in Cell Biology. https://doi.org/10.1016/j.tcb.2021.12.004
Ali, M. A., Misko, O., Salumaa, S. O., Papkov, M., Palo, K., Fishman, D., & Parts, L. (2021). Evaluating very deep convolutional neural networks for nucleus segmentation from brightfield cell microscopy images. SLAS DISCOVERY: Advancing the Science of Drug Discovery, 26(9), 1125–1137. https://doi.org/10.1177/24725552211023214
Christiansen, E. M., Yang, S. J., Ando, D. M., Javaherian, A., Skibinski, G., Lipnick, S., & Finkbeiner, S. (2018). In silico labeling: predicting fluorescent labels in unlabeled images. Cell, 173(3), 792–803. https://doi.org/10.1016/j.cell.2018.03.040
Sadanandan, S. K., Ranefall, P., Le Guyader, S., & Wählby, C. (2017). Automated training of deep convolutional neural networks for cell segmentation. Scientific Reports, 7(1), 7860. https://doi.org/10.1038/s41598-017-07599-6
Carpenter, A. E., Jones, T. R., Lamprecht, M. R., Clarke, C., Kang, I. H., Friman, O., & Sabatini, D. M. (2006). Cell profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology, 7, 1–11. https://doi.org/10.1186/gb-2006-7-10-r100
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2019). Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE transactions on medical imaging, 39(6), 1856–1867. https://doi.org/10.1109/TMI.2019.2959609
Ren, H., Zhao, M., Liu, B., Yao, R., Liu, Q., Ren, Z., & Tang, C. (2020). Cellbow: a robust customizable cell segmentation program. Quantitative Biology, 8(3), 245–255. https://doi.org/10.1007/s40484-020-0213-6
**ao, H., Li, L., Liu, Q., Zhu, X., & Zhang, Q. (2023). Transformers in medical image segmentation: A review. Biomedical Signal Processing and Control, 84, 104791. https://doi.org/10.1016/j.bspc.2023.104791
Thisanke, H., Deshan, C., Chamith, K., Seneviratne, S., Vidanaarachchi, R., & Herath, D. (2023). Semantic segmentation using vision transformers: A survey. Engineering Applications of Artificial Intelligence, 126, 106669. https://doi.org/10.1016/j.engappai.2023.106669
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., ... & Guo, B. (2021). Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF international conference on computer vision (pp. 10012–10022). https://doi.org/10.48550/ar**v.2103.14030
Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., & Wang, M. (2022, October). Swin-unet: Unet-like pure transformer for medical image segmentation. In European conference on computer vision (pp. 205–218). Cham: Springer Nature Switzerland.
Chandrasekaran, S. N., Cimini, B. A., Goodale, A., Miller, L., Kost-Alimova, M., Jamali, N., & Carpenter, A. E. (2024). Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations. Nature Methods. https://doi.org/10.1038/s41592-024-02241-6
Singh, S., Bray, M. A., Jones, T. R., & Carpenter, A. E. (2014). Pipeline for illumination correction of images for high-throughput microscopy. Journal of microscopy, 256(3), 231–236. https://doi.org/10.1111/jmi.12178
Bray, M. A., Gustafsdottir, S. M., Rohban, M. H., Singh, S., Ljosa, V., Sokolnicki, K. L., & Carpenter, A. E. (2017). A dataset of images and morphological profiles of 30 000 small-molecule treatments using the cell painting assay. Gigascience, 6(12), giw014. https://doi.org/10.1093/gigascience/giw014
Sultana, F., Sufian, A., & Dutta, P. (2020). Evolution of image segmentation using deep convolutional neural network: A survey. Knowledge-Based Systems, 201, 106062. https://doi.org/10.1016/j.knosys.2020.106062
Lin, S., & Norouzi, N. (2021). An effective deep learning framework for cell segmentation in microscopy images. In 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (pp. 3201–3204). IEEE. https://doi.org/10.1109/EMBC46164.2021.9629863
Jena, B., Digdarshi, D., Paul, S., Nayak, G. K., & Saxena, S. (2023). Effect of learning parameters on the performance of the U-Net architecture for cell nuclei segmentation from microscopic cell images. Microscopy, 72(3), 249–264. https://doi.org/10.1093/jmicro/dfac063
Zaheer, R., & Shaziya, H. (2019). A study of the optimization algorithms in deep learning. In 2019 third international conference on inventive systems and control (ICISC) (pp. 536–539). IEEE. https://doi.org/10.1109/ICISC44355.2019.9036442
Lin, A., Chen, B., Xu, J., Zhang, Z., Lu, G., & Zhang, D. (2022). Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Transactions on Instrumentation and Measurement, 71, 1–15. https://doi.org/10.1109/TIM.2022.3178991
Al Qurri, A., & Almekkawy, M. (2023). Improved UNet with attention for medical image segmentation. Sensors, 23(20), 8589. https://doi.org/10.3390/s23208589
Schabath, M. B., & Cote, M. L. (2019). Cancer progress and priorities: Lung cancer. Cancer epidemiology, biomarkers & prevention, 28(10), 1563–1579. https://doi.org/10.1158/1055-9965.EPI-19-0221
Huang, H. C., Chiang, S. J., Wen, S. H., Lee, P. J., Chen, H. W., Chen, Y. F., & Dong, C. Y. (2019). Three-dimensional nucleus-to-cytoplasm ratios provide better discrimination of normal and lung adenocarcinoma cells than in two dimensions. Journal of Biomedical Optics, 24(8), 080502–080502. https://doi.org/10.1117/1.JBO.24.8.080502
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Sreelekshmi Palliyil Sreekumar: Methodology, Writing – original draft; Rohini Palanisamy: Methodology, Writing – review & editing; Ramakrishnan Swaminathan: Conceptualization, Writing – review & editing, Supervision.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical Approval and Consent to Participate
As this study utilizes publicly available dataset, ethics approval and consent to participate are not applicable.
Consent for Publication
The authors provide full consent for submission and publishing the manuscript in the journal.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sreekumar, S.P., Palanisamy, R. & Swaminathan, R. An Approach to Segment Nuclei and Cytoplasm in Lung Cancer Brightfield Images Using Hybrid Swin-Unet Transformer. J. Med. Biol. Eng. 44, 448–459 (2024). https://doi.org/10.1007/s40846-024-00873-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-024-00873-9