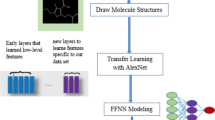
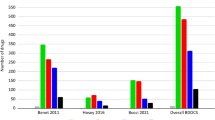
Abstract
Background and Objective
The biopharmaceutics drug disposition classification system (BDDCS) categorizes drugs into four classes on the basis of their solubility and metabolism. This framework allows for the study of the pharmacokinetics of transporters and enzymatic metabolization on biopharmaceuticals, as well as drug–drug interactions in the body. The objective of the present study was to develop computational models by neural network models and structural parameters and physicochemical properties to estimate the class of a drug in the BDDCS system.
Methods
In this study, deep learning methods were utilized to explore the potential of artificial and convolutional neural networks (ANNs and CNNs) in predicting the BDDCS class of 721 substances. The structural parameters and physicochemical properties [Abraham solvation parameters, octanol-water partition (log P) and over the pH range 1–7.5 (log D), number of rotatable bonds, hydrogen bond acceptor numbers, as well as hydrogen bond donor count] are calculated with various software. These compounds were then split into a training set consisting of 602 molecules and a test set of 119 compounds to validate the models.
Results
The results of this study showed that neural network models using applied parameters of the drug, i.e., log D and Abraham solvation parameters, are able to predict the class of solubility and metabolism in the BDDCS system with good accuracy.
Conclusions
Neural network models are well equipped to deal with the relations between the structural parameters and physicochemical properties of drugs and BDDCS classes. In addition, log D is a more suitable parameter compared with log P in predicting BDDCS.



Similar content being viewed by others
References
Wu CY, Benet LZ. Predicting drug disposition via application of BCS: Transport/absorption/ elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm Res. 2005;22(1):11–23. https://doi.org/10.1007/s11095-004-9004-4.
Bocci G, Benet LZ, Oprea TI. Can BDDCS illuminate targets in drug design? Drug Discov Today. 2019;24(12):2299–306. https://doi.org/10.1016/j.drudis.2019.09.021.
Charalabidis A, Sfouni M, Bergström C, Macheras P. The biopharmaceutics classification system (BCS) and the biopharmaceutics drug disposition classification system (BDDCS): beyond guidelines. Int J Pharm. 2019;566:264–81. https://doi.org/10.1016/j.ijpharm.2019.05.041.
Abdolmaleki A, Ghasemi JB, Ghasemi F. Computer aided drug design for multi-target drug design: SAR /QSAR, molecular docking and pharmacophore methods. Curr Drug Targets. 2017;18(5):556–75. https://doi.org/10.2174/1389450117666160101120822.
Patel V, Shah M. Artificial intelligence and machine learning in drug discovery and development. Intell Med. 2022;2(3):134–40. https://doi.org/10.1016/j.imed.2021.10.001.
Vatansever S, Schlessinger A, Wacker D, Kaniskan HÜ, ** J, Zhou MM, Zhang B. Artificial intelligence and machine learning-aided drug discovery in central nervous system diseases: state-of-the-arts and future directions. Med Res Rev. 2021;41(3):1427–73. https://doi.org/10.1002/med.21764.
Hanafi A, Amani A. Effect of processing/formulation parameters on particle size of nanoemulsions containing ibuprofen: an artificial neural networks study. Pharm Sci. 2021;27(2):230–7. https://doi.org/10.34172/PS.2020.74.
Goodfellow I, Bengio Y, Courville A. Deep learning. MIT Press; 2016.
Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611–29. https://doi.org/10.1007/s13244-018-0639-9.
Golfar Y, Shayanfar A. Prediction of biopharmaceutical drug disposition classification system (BDDCS) by structural parameters. J Pharm Pharm Sci. 2019;22(1):247–69. https://doi.org/10.18433/jpps30271.
Broccatelli F, Cruciani G, Benet LZ, Oprea TI. Bddcs class prediction for new molecular entities. Mol Pharm. 2012;9(3):570–80. https://doi.org/10.1021/mp2004302.
Lobo S. Is there enough focus on lipophilicity in drug discovery? Expert Opin Drug Discov. 2020;15(3):261–3. https://doi.org/10.1080/17460441.2020.1691995.
Ghotbi G, Hamzeh-Mivehroud M, Taghvimi A, Davaran S, Dastmalchi S. Investigation of experimental and in silico physicochemical properties of thiazole-pyridinium anti-acetylcholinesterase derivatives with potential anti-Alzheimer’s activity. Pharm Sci. 2021;27(3):366–77. https://doi.org/10.34172/PS.2020.81.
Papich MG, Martinez MN. Applying Biopharmaceutical Classification System (BCS) criteria to predict oral absorption of drugs in dogs: challenges and pitfalls. AAPS J. 2015;17(4):948–64. https://doi.org/10.1208/s12248-015-9743-7.
Jouyban A, Acree WE, Michael H Jr. Abraham and his developed parameters: various applications in medicine, chemistry and biology. Pharm Sci. 2022;28(2):170–3. https://doi.org/10.34172/PS.2022.1.
Aliagas I, Gobbi A, Lee ML, Sellers BD. Comparison of log P and log D correction models trained with public and proprietary data sets. J Comp-Aided Mol Des. 2022;36(3):253–62. https://doi.org/10.1007/s10822-022-00450-9.
Benet LZ, Broccatelli F, Oprea TI. BDDCS applied to over 900 drugs. AAPS J. 2011;13(4):519–47. https://doi.org/10.1208/s12248-011-9290-9.
Hosey CM, Chan R, Benet LZ. BDDCS predictions, self-correcting aspects of BDDCS assignments, BDDCS assignment corrections, and classification for more than 175 additional drugs. AAPS J. 2016;18(1):251–60. https://doi.org/10.1208/s12248-015-9845-2.
Bocci G, Oprea TI, Benet LZ. State of the art and uses for the biopharmaceutics drug disposition classification system (BDDCS): new additions, revisions, and citation references. AAPS J. 2022. https://doi.org/10.1208/s12248-022-00687-0.
Ulrich N, Endo S, Brown TN, Watanabe N, Bronner G, Abraham MH, Goss KU, UFZ-LSER database v 3.2.1 [Internet], Leipzig, Germany, Helmholtz Centre for Environmental Research-UFZ. 2017. http://www.ufz.de/lserd. Accessed on Aug 2022.
Reaxys, https://www.reaxys.com/. Accessed on Aug 2022.
ACD/labs, https://ilab.acdlabs.com. Accessed on August 2022.
Siramshetty V, Williams J, Nguyễn ÐT, Neyra J, Southall N, Mathé E, Xu X, Shah P. Validating ADME QSAR models using marketed drugs. SLAS Discov. 2021;26(10):1326–36. https://doi.org/10.1177/24725552211017520.
Mohammadi SM, Shayanfar A, Emami S, Jouyban A. Effects of amount of excess solid, the type of stirring and sedimentation time on solubility of sodium phenytoin and lamotrigine. ADMET DMPK. 2018;6(4):269–78. https://doi.org/10.5599/admet.621.
Tinworth CP, Young RJ. Facts, patterns, and principles in drug discovery: appraising the rule of 5 with measured physicochemical data. J Med Chem. 2020;63(18):10091–108. https://doi.org/10.1021/acs.jmedchem.9b01596.
Kah M, Brown CD. Log D: lipophilicity for ionisable compounds. Chemosphere. 2008;72(10):1401–8. https://doi.org/10.1016/j.chemosphere.2008.04.074.
Benet LZ. The role of BCS (biopharmaceutics classification system) and BDDCS (biopharmaceutics drug disposition classification system) in drug development. J Pharm Sci. 2013;102(1):34–42. https://doi.org/10.1002/jps.23359.
Bhal SK, Kassam K, Peirson IG, Pearl GM. The rule of five revisited: applying log D in place of log P in drug-likeness filters. Mol Pharm. 2007;4(4):556–60. https://doi.org/10.1021/mp0700209.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
A.S. would like to acknowledge financial support from the Vice Chancellor for Research of Tabriz University of Medical Sciences, Tabriz, Iran (no.: 66944).
Conflict of Interest
Aryan Ashrafi, Kiarash Teymouri, Farnaz Aghazadeh, and Ali Shayanfar have no conflicts of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Availability of Data
All data are available as a supplementary file (Table S1) on the journal’s website along with the published article.
Code Availability
It is available online in the following link: https://github.com/AryanAshrafi/Neural-Network-models-for-predicting-BDDCS-system.
Author Contributions
Aryan Ashrafi was responsible for data analysis and interpretation and drafting the article; Kiarash Teymouri for data analysis and interpretation and drafting the article; Farnaz Aghazadeh for data collection; and Ali Shayanfar for design of the work, supervision of the project, and critical revision of the article. All authors read and approved the final manuscript.
Consent to Publish
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ashrafi, A., Teimouri, K., Aghazadeh, F. et al. Neural Network Models for Predicting Solubility and Metabolism Class of Drugs in the Biopharmaceutics Drug Disposition Classification System (BDDCS). Eur J Drug Metab Pharmacokinet 49, 1–6 (2024). https://doi.org/10.1007/s13318-023-00861-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13318-023-00861-5