Abstract
Background
The pandemic 2009 swine flu is a highly infectious respiratory disorder caused by H1N1 influenza A viruses. A recent study reported that knockout of the prion protein gene (PRNP) induced susceptibility and lethality in influenza A virus-infected mice.
Objective
Thus, we examined the association between genetic variations of the PRNP gene and susceptibility to pandemic 2009 swine flu.
Results
We did not find an association between PRNP polymorphisms and susceptibility to pandemic 2009 swine flu.
Conclusions
To the best of our knowledge, this was the first evaluation of the association between PRNP polymorphisms and vulnerability to pandemic 2009 swine flu.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Influenza A viruses are negative-sense single-stranded RNA viruses causing respiratory disorders in humans and animals. Since the genome of influenza A viruses is segmented RNA, the genome can be reassorted through the life cycle of various types of influenza A viruses and cause antigenic shifts (Javanian et al. 2021; Moghadami 2017). The pandemic influenza A H1N1 2009 virus is caused by an antigenic shift mediated by quadruple reassortments of viral genomes, including swine, avian, and human influenza A viruses (Allen et al. 2017; Everitt et al. 2012; Girard et al. 2010; Kim et al. 2020; Schnitzler and Schnitzler 2009). In response to the invasion of influenza A viruses, macrophages are activated to clear pathogens. However, macrophages are also the major reservoir of reactive oxygen species (ROS). The generation of excessive ROS results in the destruction of epithelial cell layers, which are considered the first line of antiviral defense for the host (Lin et al. 2016; Reshi et al. 2014; Kim et al. 2021a, b).
The prion protein (PrP), encoded by the prion protein gene (PRNP), is a glycosylphosphatidylinositol (GPI)-anchored multifunctional protein composed of a nonstructural octapeptide repeat domain and a globular C-terminal domain (Kim et al. 2021c; Prusiner 1998a, b). Previous studies have reported that PrP plays a pivotal role in the protection of oxidative stress in several organs, including the brain, heart and lungs (Castle and Gill 2017). In addition, deficiency of the PRNP gene confers susceptibility and lethality in influenza A virus-infected mice (Chida et al. 2018). Thus, we postulated that genetic variations in the PRNP gene, which are related to the function and expression level of PrP, may be associated with susceptibility to influenza A viruses.
To investigate the association between PRNP polymorphisms and vulnerability to pandemic 2009 swine flu, we investigated the genotype and allele frequencies of the PRNP polymorphisms located on the open reading frame (ORF) and transcriptional regulatory region in 97 healthy control subjects and 30 pandemic 2009 swine flu-infected patients using direct sequencing. We evaluated an association between PRNP polymorphisms and susceptibility to pandemic 2009 swine flu infection by comparing the genotype and allele frequencies of the PRNP polymorphisms between these two groups.
Materials and methods
Ethics statements
All samples were collected with informed consent under institutional review board-approved protocols. All experimental procedures were approved following the guidelines of the institutional review board (IRB) of Jeonbuk National University and the 1964 Helsinki Declaration and its later amendments (approval number: JBNU 2017-08-009). All the samples and information were anonymized prior to study.
Subject
Detailed information on all subjects was explained in a previous study (Kim et al. 2020). In brief, healthy controls and pandemic 2009 swine flu-infected patients have no underlying disease and co-morbidity (Table 1).
Genomic DNA extraction
Genomic DNA was isolated from 200 μl of peripheral blood using the Blood Genomic DNA Isolation Kit (Qiagen, Valencia, California, USA) following the manufacturer’s protocol.
Genetic analysis
The PRNP gene was amplified by polymerase chain reaction (PCR) using gene-targeted primers. Detailed information on the primers and experimental conditions are described in Table 2. PCR was carried out using GoTaq® DNA Polymerase (Promega, Fitchburg, Wisconsin, USA) and an S-1000 Thermal Cycler (Bio–Rad, Hercules, California, USA) following the manufacturer’s protocol. The PCR products were directly analyzed with an ABI 3730 automatic sequencer (ABI, Foster City, California, USA) and the results were annotated by Finch TV software (Geospiza Inc, Seattle, USA).
Statistical analysis
Statistical analyses were performed using SAS version 9.4 (SAS Institute Inc., USA).
Results
Investigation of polymorphisms of the PRNP gene
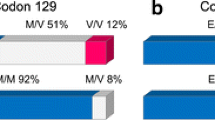
To investigate the genotype and allele frequencies of PRNP polymorphisms in the Korean population, we performed direct sequencing in 97 healthy controls and 30 pandemic 2009 swine flu-infected patients. The sequenced products were homologous to the PRNP gene of Homo sapiens registered in GenBank (Gene ID: 5621). We performed genoty** for 2 SNPs, M129V and E219K, located on the ORF of the PRNP gene (Table 3), and 3 SNPs, c.1368 T > C, c.1380 T > C and c.1424G > A, located on the transcriptional regulatory region of the PRNP gene (Table 4).
Evaluation of an association between PRNP polymorphisms and vulnerability to pandemic 2009 swine flu
To examine an association between the genetic distribution of the PRNP gene and susceptibility to pandemic 2009 swine flu infection, we compared the genotype and allele frequencies of the PRNP gene polymorphisms between the healthy control subjects and pandemic 2009 swine flu-affected patients.
Interestingly, there is no association of the genotype and allele distributions of 2 SNPs, M129V and E219K, of the PRNP gene located on the ORF with vulnerability to pandemic 2009 swine flu (Table 3). In addition, there is no association of the genotype and allele distributions of 3 SNPs, c.1368 T > C, c.1380 T > C and c.1424G > A, located in transcriptional regulatory region of the PRNP gene with susceptibility to pandemic 2009 swine flu (Table 4).
Discussion
Previous studies have reported that nonsynonymous genetic polymorphisms of the PRNP gene modulate structural alterations of PrP and are related to susceptibility to several types of prion diseases, including Creutzfeldt–Jakob disease (CJD) in humans and chronic wasting disease (CWD) in elks and deer. In humans, genetic variations of the human PRNP gene at codons 129 and 219 play a major role in susceptibility to CJD (Alperovitch et al. 1999; Jeong et al. 2005; Lee et al. 2001; Lloyd et al. 2011; Vollmert et al. 2006). In addition, nonsynonymous SNPs of the cervid PRNP gene at codons 95, 96, and 132 are also associated with vulnerability to CWD (Arifin et al. 2021; Johnson et al. 2006; Robinson et al. 2012). However, we did not find an association between functional genetic variations, including M129V and E219K, and vulnerability to pandemic 2009 swine flu (Table 3). In addition, previous studies have reported that genetic polymorphisms found in the promoter of the PRNP gene are involved in susceptibility to CJD and bovine spongiform encephalopathy (BSE). In humans, c.1368 T > C (rs1029273), located in the promoter region plays a pivotal role in susceptibility to sporadic CJD in British and German populations (Bratosiewicz-Wasik et al. 2012; Mastrianni 2010; Mead et al. 2001). In addition, 23- and 12-bp insertion/deletion polymorphisms located in the transcription regulatory region of the bovine PRNP gene are related to the expression level of the bovine PRNP gene and affect vulnerability to BSE (Haase et al. 2007; Murdoch and Murdoch 2015). Notably, we found no association between promoter polymorphisms of the PRNP gene and vulnerability to the pandemic 2009 swine flu in the present study (Table 4). It indicates that PRNP polymorphisms do not affect the infection mechanism of the pandemic 2009 swine flu. However, since this evaluation has been performed in relatively small cohorts, further confirmation in large cohorts would be highly advantageous in the future. In addition, Koreans have highly heterogeneous genetic background of the PRNP gene compared to other ethnic groups, and further investigation of the association analysis in other ethnic groups is needed in the future. In the present study, only one patient was admitted to the ICU (Table 1). Thus, we could not perform the association analysis between the disease severity and the PRNP polymorphisms. Since the PrP is related to lethality in influenza A virus-infected mice, PRNP polymorphisms may be related to the disease severity. Thus, further association analysis stratified by severity, including the entrance of the intensive care unit and/or pneumonia, is needed to validate the association between entire PRNP polymorphisms and the clinical outcome of the pandemic 2009 swine flu-infected patients (Chida et al. 2018).
In a recent study, the MX1 protein, which showed potent antiviral activity, played a pivotal role in blocking H7N9 influenza viruses (Chen et al. 2021). Furthermore, genetic variations of the MX1 gene are associated with susceptibility to influenza viruses. Since H7N9 influenza viruses showed a similar pathomechanism to H1N1 influenza viruses, further investigation between MX1 polymorphisms and susceptibility to pandemic 2009 swine flu is highly desirable in the future.
Conclusions
In this study, we investigated genetic variations of the PRNP gene in healthy control subjects and the pandemic 2009 swine flu-infected patients using direct sequencing. We evaluated an association between PRNP polymorphisms and susceptibility to the pandemic 2009 swine flu infection. However, we did not find an association between these polymorphisms and susceptibility to pandemic 2009 swine flu.
Data availability statement
All data are available from the corresponding authors upon reasonable request.
References
Allen EK et al (2017) SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat Med 23:975–983
Alperovitch A et al (1999) Codon 129 prion protein genotype and sporadic Creutzfeldt–Jakob disease. Lancet 353:1673–1674
Arifin MI et al (2021) Cervid prion protein polymorphisms: role in chronic wasting disease pathogenesis. Int J Mol Sci 22:2271
Bratosiewicz-Wasik J et al (2012) Association between the PRNP 1368 polymorphism and the occurrence of sporadic Creutzfeldt–Jakob disease. Prion 6:413–416
Castle AR, Gill AC (2017) Physiological functions of the cellular prion protein. Front Mol Biosci 4:19
Chen Y et al (2021) Rare variant MX1 alleles increase human susceptibility to zoonotic H7N9 influenza virus. Science 373:918–922
Chida J et al (2018) Prion protein protects mice from lethal infection with influenza A viruses. PLoS Pathog 14:e1007049
Everitt AR et al (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484:519–523
Girard MP et al (2010) The 2009 A (H1N1) influenza virus pandemic: a review. Vaccine 28:4895–4902
Haase B et al (2007) PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC Genet 8:15
Javanian M et al (2021) A brief review of influenza virus infection. J Med Virol 93:4638–4646
Jeong BH et al (2005) Association of sporadic Creutzfeldt–Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics 6:229–232
Johnson C et al (2006) Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol 87:2109–2114
Kim YC, Jeong MJ, Jeong BH (2020) Strong association of regulatory single nucleotide polymorphisms (SNPs) of the IFITM3 gene with influenza H1N1 2009 pandemic virus infection. Cell Mol Immunol 17:662–664
Kim YC, Jeong MJ, Jeong BH (2021a) Genetic association between the rs12252 SNP of the interferon-induced transmembrane protein gene and influenza A virus infection in the Korean population. Mol Cell Toxicol 17:51–57
Kim YC, Won SY, Jeong BH (2021b) The first association study of single-nucleotide polymorphisms (SNPs) of the IFITM1 gene with influenza H1N1 2009 pandemic virus infection. Mol Cell Toxicol 17:179–186
Kim YC, Won SY, Jeong BH (2021c) Altered expression of glymphatic system-related proteins in prion diseases: Implications for the role of the glymphatic system in prion diseases. Cell Mol Immunol 18:2281–2283
Lee HS et al (2001) Increased susceptibility to Kuru of carriers of the PRNP 129 methionine/methionine genotype. J Infect Dis 183:192–196
Lin X et al (2016) The influenza virus H5N1 infection can induce ROS production for viral replication and host cell death in A549 cells modulated by human Cu/Zn superoxide dismutase (SOD1) overexpression. Viruses 8:13
Lloyd S, Mead S, Collinge J (2011) Genetics of prion disease. Top Curr Chem 305:1–22
Mastrianni JA (2010) The genetics of prion diseases. Genet Med 12:187–195
Mead S et al (2001) Sporadic–but not variant–Creutzfeldt-Jakob disease is associated with polymorphisms upstream of PRNP exon 1. Am J Hum Genet 69:1225–1235
Moghadami M (2017) A narrative review of influenza: a seasonal and pandemic disease. Iran J Med Sci 42:2–13
Murdoch BM, Murdoch GK (2015) Genetics of prion disease in cattle. Bioinform Biol Insights 9:1–10
Prusiner SB (1998a) The prion diseases. Brain Pathol 8:499–513
Prusiner SB (1998b) Prions. Proc Natl Acad Sci U S A 95:13363–13383
Reshi ML, Su YC, Hong JR (2014) RNA viruses: ROS-mediated cell death. Int J Cell Biol 2014:467452
Robinson SJ et al (2012) The role of genetics in chronic wasting disease of North American cervids. Prion 6:153–162
Schnitzler SU, Schnitzler P (2009) An update on swine-origin influenza virus A/H1N1: a review. Virus Genes 39:279–292
Vollmert C et al (2006) Significant association of a M129V independent polymorphism in the 5′ UTR of the PRNP gene with sporadic Creutzfeldt-Jakob disease in a large German case-control study. J Med Genet 43:e53
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2021R1A2C1013213, 2022R1C1C2004792). This work was supported by NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program). Yong-Chan Kim was supported by the BK21 Plus Program in the Department of Bioactive Material Sciences. This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876, 2021R1A6A3A01086488).
Author information
Authors and Affiliations
Contributions
YCK, SYW and BHJ conceived and designed the experiment. YCK and SYW performed the experiments. YCK, SYW and BHJ analyzed the data. YCK and BHJ wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Yong-Chan Kim declares that he/she has no conflict of interest. Sae-Young Won declares that he/she has no conflict of interest. Byung-Hoon Jeong declares that he/she has no conflict of interest.
Ethical approval
All samples were collected with informed consent under institutional review board-approved protocols. All experimental procedures were approved following the guidelines of the institutional review board (IRB) of Jeonbuk National University and the 1964 Helsinki Declaration and its later amendments (approval number: JBNU 2017-08-009). All the samples and information were anonymized prior to study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, YC., Won, SY. & Jeong, BH. No association of prion protein gene (PRNP) polymorphisms with susceptibility to the pandemic 2009 swine flu. Mol. Cell. Toxicol. 19, 799–803 (2023). https://doi.org/10.1007/s13273-022-00318-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13273-022-00318-x