Abstract
Purpose
The Gleason score (GS) and positive needles are crucial aggressive indicators of prostate cancer (PCa). This study aimed to investigate the usefulness of magnetic resonance imaging (MRI) radiomics models in predicting GS and positive needles of systematic biopsy in PCa.
Material and Methods
A total of 218 patients with pathologically proven PCa were retrospectively recruited from 2 centers. Small-field-of-view high-resolution T2-weighted imaging and post-contrast delayed sequences were selected to extract radiomics features. Then, analysis of variance and recursive feature elimination were applied to remove redundant features. Radiomics models for predicting GS and positive needles were constructed based on MRI and various classifiers, including support vector machine, linear discriminant analysis, logistic regression (LR), and LR using the least absolute shrinkage and selection operator. The models were evaluated with the area under the curve (AUC) of the receiver-operating characteristic.
Results
The 11 features were chosen as the primary feature subset for the GS prediction, whereas the 5 features were chosen for positive needle prediction. LR was chosen as classifier to construct the radiomics models. For GS prediction, the AUC of the radiomics models was 0.811, 0.814, and 0.717 in the training, internal validation, and external validation sets, respectively. For positive needle prediction, the AUC was 0.806, 0.811, and 0.791 in the training, internal validation, and external validation sets, respectively.
Conclusions
MRI radiomics models are suitable for predicting GS and positive needles of systematic biopsy in PCa. The models can be used to identify aggressive PCa using a noninvasive, repeatable, and accurate diagnostic method.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Prostate cancer (PCa) has the highest incidence rate and the second-highest mortality rate in the male reproductive system worldwide [1]. Aggressive PCas have been proven to be a critical factor in the selection of clinical treatment strategies and assessment of prognosis. Patients with low-to-moderate aggressive PCas have satisfactory 5- and 10- year survival rates compared with those with highly aggressive PCas [2].
The Gleason score (GS) is the most widely used tool to assess the aggressiveness of PCa [3,4,5]. Studies have reported that the number of positive needles of systematic biopsy is also one of the indicators for assessing PCa aggressiveness [6]. A considerable difference in prognosis exists between patients with ≥ 50% positive needles and those with < 50% positive needles [7]. No study has yet reported to predict the number of positive needles.
Although magnetic resonance imaging (MRI) has a fair sensitivity for detecting and localizing PCa, certain small-volume lesions may be missed; thus, a relatively higher image quality and expertise of radiologists are required for accurate diagnosis [8, 9]. Radiomics is currently regarded as a promising noninvasive tool to characterize PCa aggressiveness. Previous studies [3, 10] have demonstrated that the machine learning model constructed by extracting MRI-based radiomics features from the whole prostate has satisfactory performance in predicting GS, and the area surrounding the tumor lesion provides information related to GS. Therefore, in this study, we aimed to develop radiomics models based on prostate gland segmentation to predict PCa aggressiveness regarding GS and positive needles of systematic biopsy.
2 Materials and methods
2.1 Patients
This study received ethics approval from the Medical Ethics Committee of Gansu Provincial Hospital (2022–458), and all patients were exempted from signing an informed consent form. Furthermore, all methods employed in this study were performed in strict accordance with the relevant guidelines and regulations around a local institution’s radiomics model.
The clinicopathologic data on patients with PCa were collected retrospectively from the electronic medical systems of 2 centers (Center A, Gansu Provincial Hospital from January 2018 to July 2023; Center B, Zhangye People's Hospital Affiliated to Hexi University from April 2020 to August 2023).
The clinical data, including age, serum total prostate-specific antigen (TPSA) level, serum-free prostate-specific antigen (FPSA) level, FPSA/TPSA, alkaline phosphatase (ALP) level, GS, and positive needle biopsy. GS and positive needle biopsy were obtained from medical records and pathologic reports. In this study, PSA density (PSAD) was calculated using the following formula: PSAD = TPSA/VPG, where VPG is the volume of the whole prostate gland. The value of VPG was obtained using small-field-of-view high-resolution T2-weighted imaging (sFOV HR-T2WI).
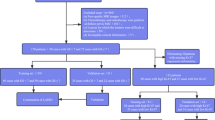
The inclusion criteria were as follows: (a) pathologically confirmed PCa by systematic biopsy; (b) prostate biopsy performed within 1 month after MRI examination; and (c) no prostate therapy, such as surgery, androgen deprivation therapy, or radiotherapy, administered prior to MRI examination. The exclusion criteria were as follows: (a) clinical, pathologic, or MRI imaging data missing; (b) systemic puncture without 12-needle biopsy; (c) poor MRI image quality; and (d) presence of other tumors. The general flowchart of this study is depicted in Fig. 1.
2.2 MRI acquisition
All enrolled patients underwent pelvic MRI examination on a 3.0-T MRI scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany; Siemens lumina 214,707) with the 18-channel phased-array abdominal coil. The center of the coil was aligned just above the pubic symphysis, and the coil was secured using an abdominal strap to minimize the influence of respiratory motion artifacts in the abdomen. The set image protocol included sagittal turbo spin-echo–T2WI and axial fat-suppressed–T2WI, axial diffusion weighted imaging (DWI) (b values of 50 and 1000) with apparent diffusion coefficient (ADC) maps, sFOV high-resolution turbo T2-weighted sequences on the axial planes, and post-contrast delayed scan sequences. The MRI data sets were retrieved from the picture achieving and communication system for further image processing. The MRI scan parameters are detailed in Table S1.
2.3 The method of systematic biopsy for PCa
All patients were diagnosed of PCa by ultrasound guided transrectal prostate biopsy using the standard 10 + X puncture method (1 ~ 3 cores). The biopsy specimens were sent to the pathology department for routine histopathological examination, and the results were interpreted and recorded by a professional pathologist specializing in the urinary system.
2.4 Region of interest segmentation
The sFOV HR-T2WI and post-contrast delayed scan images were imported into the open-source ITK-SNAP software (version 3.8.0, www.itksnap.org) in Digital Imaging and Communications in Medicine (DICOM) format. A radiologist with 5 years of experience in diagnosing prostate diseases routinely performed manual segmentation of the region of interest (ROI) layer by layer on the sFOV HR-T2WI images and post-contrast delayed scan images, avoiding the patient’s urethra, ejaculatory ducts, seminal vesicles, and the base of the seminal vesicles. Considering the necessity of overall ROI analysis in the heterogeneity assessment, the ROI included areas of hemorrhage, necrosis, cystic changes, and calcifications. After a rough outlining of the prostate gland, the software automatically generated the 3-dimensional (3D) volume of interest (VOI) for the entire prostate. Figure 2 depicts an example of the 3D manual segmentation. Figure S1 shows MRI images and pathological results of two PCa patients. A random sample of 30 patients was selected, and the same radiologist performed ROI after 3 months. This was conducted to assess the reproducibility and stability of radiomics features.
2.5 Radiomics feature extraction
FeAture Explorer (FAE), which is a visualization program (version 0.5.5, https://github.com/salan668/FAE), was used for feature extraction after completing segmentation. A linear interpolation was adopted to resample the voxel size of the image to an isovolumetric voxel (1 × 1 × 1 mm3) before feature extraction. The voxel intensities of the image discretization were applied with a fixed bin width of 25 to reduce the influence of various machine types and scanning parameters on radiomics features. A series of quantitative radiomics features were extracted from the VOIs of axial sFOV HR-T2WI and post-contrast delayed scan images. The radiomics features were of 7 types: (a) first order; (b) shape-based; (c) gray-level co-occurrence matrix; (d) gray-level run-length matrix; (e) gray-level size zone matrix; (f) gray-level dependence matrix; and (g) neighboring gray-tone difference matrix.
2.6 Feature selection and radiomics model establishment
For GS prediction, we performed upsampling of both positive and negative samples to reduce the imbalance in the classification training set data. The Z-score was used to normalize the features. For positive needle prediction, we performed SMOTE of both positive and negative samples to reduce the imbalance in the classification training set data. The Minimax was used to normalize the features. Furthermore, we applied Pearson correlation coefficients (PCC) on each pair of features to decrease the dimensionality of the feature matrix. We casually removed one of them with PCC > 0.9.
The analysis of variance (ANOVA), relief algorithm, recursive feature elimination, and Kruskal–Wallis test were used to select the most optimal features with nonzero coefficients from the retained standardized features and establish the radiomics signature. The support vector machine, linear discriminant analysis, logistic regression (LR), and LR using the least absolute shrinkage and selection operator were used to obtain the optimal feature combination based on accuracy step by step. We used the tenfold cross-validation to set the parameters according to the model performance on the validation data sets.
2.7 Predictive performance of the radiomics models
The discrimination ability of the radiomics models was assessed and validated in the training, internal validation, and external validation sets using the area under the curve (AUC) of the receiver operating characteristic (ROC). Simultaneously, the accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated based on the cutoff value that maximized the Youden index. The Matthews correlation coefficient (MCC) was a scalar measure commonly used to assess classification quality. The goodness-of-fit of the radiomics models was estimated using the calibration curve and the Hosmer–Lemeshow test. The decision curve analysis (DCA) demonstrated the clinical net benefits of the radiomics models [11].
2.8 Statistical analysis
The data were statistically analyzed using the SPSS software (version 21.0, IBM, https://www.ibm.com/cn-zh/analytics/spss-statistics-software), GraphPad Prism (version 9.0.0, https://www.graphpad.com/), and R software (version 4.3.0). The Kolmogorov–Smirnov test was used to assess whether the continuous variable was normally distributed. Continuous variables conforming to a normal distribution were expressed as means ± standard deviation. The differences between 3 groups were evaluated using ANOVA and those between 2 groups using the independent-samples t test when appropriate. Qualitative and continuous variables with a nonnormal distribution were expressed as the medians (lower and upper quartiles). The differences among 3 groups were first tested using the Kruskal–Wallis test; if a considerable difference was identified, the difference between 2 groups was evaluated using the Mann–Whitney U test when appropriate. The Spearman rank correlation analysis was used to evaluate the correlation between positive needles and GS of systematic biopsy in PCa when the continuous variables were not normally distributed. A P value < 0.05 indicated a statistically significant difference.
3 Results
3.1 Clinical characteristics of patients
From Center A, 167 eligible patients were finally included in this study. They were randomly divided into training sets (n = 117) and internal validation sets (n = 50) at a ratio of 7:3 adopting the stratified sampling method. From Center B, 51 eligible patients were finally included in external validation sets (Fig. 1).
For GS prediction, the basic clinical information of patients in the training, internal validation, and external validation sets is presented in Table 1. Of the patients, 28 (23.9%), 12 (24%), and 26 (51%) in the training, internal validation, and external validation sets, respectively, had GS ≤ 7 (Fig. 1). For positive needle prediction, a comparison of the clinical features of the patients in the training, internal validation, and external validation sets is presented in Table 2. Of the patients, 31 (26.5%), 13 (26%), and 16 (11.8%) in the training, internal validation, and external validation sets, respectively, had positive needles < 6 (Fig. 1). Age, TPSA, FPSA, FPSA/TPSA, PSAD, and ALP were not statistically significantly different among the training, internal validation, and external validation sets (P > 0.05) (Table 2); however, TPSA, FPSA, FPSA/TPSA, and PSAD in the GS ≤ 7 and GS > 7 groups differed considerably (P < 0.05) (Table 3). Moreover, TPSA, FPSA, FPSA/TPSA, PSAD, and ALP in the positive needles ≥ 6 group were higher than those in the positive needles < 6 group, and the differences were statistically significant (P < 0.05) (Table 4).
3.2 Radiomics feature extraction and feature selection
The reliability of the extracted radiomics features in terms of Intraclass correlation coefficient (ICC) for all features was quantified, with mean ICC values of 0.982. The top 11 features were chosen as the prime feature subset for the GS prediction (Table S2 and Fig. 3), whereas the top 5 features were chosen for positive needle prediction (Table S3 and Fig. 4).
Optimal radiomics model (Zscore_PCC_ANOVA_LR) for GS prediction. Different colors represent different radiomics features, and the area represents the weighted contribution of each radiomics feature in the radiomics model. The feature with the highest weight for GS prediction was wavelet-LLH_gldm_LargeDependenceHighGrayLevelEmphasis. Zscore is the method of normalizing the features for GS prediction. PCC is a feature dimension reduction method for GS prediction. ANOVA is a feature selection method for GS prediction. LR is the classifier for radiomics model for GS prediction. PCC Pearson Correlation Coefficients. ANOVA analysis of variance. LR logistic regression
Optimal radiomics model (MinMax_PCC_RFE_LR) for positive needle prediction. Different colors represent different radiomics features, and the area represents the weighted contribution of each radiomics feature in the radiomics model. The feature with the highest weight for GS prediction was wavelet-LLH_firstorder_Kurtosis. MinMax is the method of normalizing the features for positive needle prediction. PCC is a feature dimension reduction method for positive needle prediction. RFE is a feature selection method for positive needle prediction. LR is the classifier for radiomics model for positive needle prediction. PCC Pearson correlation coefficients. RFE Recursive feature elimination. LR logistic regression
3.3 GS and positive needle prediction models
The GS and positive needles exhibited a moderate positive correlation (r = 0.415, P < 0.01). The results demonstrated that the leading model for predicting GS and positive needles was LR. For GS prediction, the AUC of the radiomics models was 0.811, 0.814, and 0.717 in the training, internal validation, and external validation sets, respectively (Table S4 and Fig. 5a). The radiomics models had an accuracy of 74%, 74%, and 69%; a sensitivity of 75%, 92%, and 54%; a specificity of 73%, 68%, and 84%; a PPV of 47%, 48%, and 78%; an NPV of 90%, 96%, and 64%; and an MCC of 42%, 51%, and 40% in the training, internal validation, and external validation sets, respectively (Table S4).
For positive needle prediction models, the AUC was 0.806, 0.811, and 0.791 in the training, internal validation, and external validation sets, respectively (Table S5; Fig. 5b). The radiomics models had an accuracy of 79%, 76%, and 76%; a sensitivity of 81%, 85%, and 75%; a specificity of 79%, 73%, and 77%; a PPV of 58%, 52%, and 60%; an NPV of 92%, 93%, and 87%; and an MCC of 55%, 51%, and 50% in the training, internal validation, and external validation sets, respectively (Table S5).
3.4 Performance assessment of the radiomics models
The calibration curve and Hosmer–Lemeshow test statistic exhibited favorable calibration in the training sets, which was confirmed in the internal and external validation sets (Fig. S2, S3a). Subsequently, the DCA was performed to analyze the clinical practicability of the radiomics models. The patients would benefit more from the radiomics models than either the treatment of all schemes or the no-treatment regimen (Fig. S2, S3b). Furthermore, the prediction efficacy was compared by ROC analyses in the training, internal validation, and external validation sets (Fig. 5).
4 Discussion
Currently, tissue biopsy remains the gold standard for PCa diagnosis. Pathologic biopsy is also a common method for estimating PCa aggressiveness, but it has various complications limiting its clinical use. Furthermore, this evaluation typically relies on a solitary biopsy of a potentially heterogeneous tumor, which can merely capture a snapshot of its biological characteristics [12]. In the present dual-center retrospective study, we built and tested radiomics models for predicting PCa aggressiveness regarding GS and positive needles of systematic biopsy. The models were validated using internal and external validation sets. The established radiomics models provided a quantifiable and individualized tool for predicting PCa aggressiveness, thus hel** in clinical decision-making.
Serum PSA is a specific marker of PCa and the only tumor marker with organ specificity. In this study, TPSA, FPSA, FPSA/TPSA, and PSAD in the GS ≤ 7 and GS > 7 groups differed considerably. It may be due to the biological differences between tumors with different grades. With the increase of serum PSA related indicators, GS will also increase. Moreover, TPSA, FPSA, FPSA/TPSA, and PSAD in the positive needles ≥ 6 group were higher than those in the positive needles < 6 group. We guessed that the number of positive needles is an indicator of the extent and localization of cancerous tissue of PCa. A higher number of positive needles (≥ 6) suggests a more extensive disease involvement.
Patient management for PCa requires an accurate evaluation of potential tumor aggressiveness [13, The data that support the findings of this study are available on request from the corresponding author. Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. Wright JL, Salinas CA, Lin DW, et al. Prostate cancer specific mortality and Gleason 7 disease differences in prostate cancer outcomes between cases with Gleason 4 + 3 and Gleason 3 + 4 tumors in a population based cohort. J Urol. 2009;182:2702–7. Rodrigues A, Santinha J, Galvão B, et al. Prediction of prostate cancer disease aggressiveness using bi-parametric MRI radiomics. Cancer. 2021. https://doi.org/10.3390/cancers13236065. Bertelli E, Mercatelli L, Marzi C, et al. Machine and deep learning prediction of prostate cancer aggressiveness using multiparametric MRI. Front Oncol. 2021;11: 802964. Hurwitz LM, Agalliu I, Albanes D, et al. Recommended definitions of aggressive prostate cancer for etiologic epidemiologic research. J Natl Cancer Inst. 2021;113:727–34. Scimeca M, Montanaro M, Bonfiglio R, et al. The ETS homologous factor (EHF) represents a useful immunohistochemical marker for predicting prostate cancer metastasis. Diagnostics. 2022. https://doi.org/10.3390/diagnostics12040800. Yang Y. Comments on National guidelines for diagnosis and treatment of prostate cancer 2022 in China (English version). Chinese J Cancer Res. 2022;34:456–7. de Rooij M, Israël B, Tummers M, et al. ESUR/ESUI consensus statements on multi-parametric MRI for the detection of clinically significant prostate cancer: quality requirements for image acquisition, interpretation and radiologists’ training. Eur Radiol. 2020;30:5404–16. Popiţa C, Popiţa AR, Andrei A, et al. Local staging of prostate cancer with multiparametric-MRI: accuracy and inter-reader agreement. Med Pharm Rep. 2020;93:150–61. Gong L, Xu M, Fang M, et al. The potential of prostate gland radiomic features in identifying the Gleason score. Comput Biol Med. 2022;144: 105318. Püllen L, Radtke JP, Wiesenfarth M, et al. External validation of novel magnetic resonance imaging-based models for prostate cancer prediction. BJU Int. 2020;125:407–16. Ramtohul T, Djerroudi L, Lissavalid E, et al. Multiparametric MRI and radiomics for the prediction of HER2-zero, -low, and -positive breast cancers. Radiology. 2023;308: e222646. Mayer R, Simone CB 2nd, Turkbey B, et al. Prostate tumor eccentricity predicts Gleason score better than prostate tumor volume. Quant Imaging Med Surg. 2022;12:1096–108. Jeon J, Olkhov-Mitsel E, **e H, et al. Temporal stability and prognostic biomarker potential of the prostate cancer urine miRNA transcriptome. J Natl Cancer Inst. 2020;112:247–55. Gong L, Xu M, Fang M, et al. Noninvasive prediction of high-grade prostate cancer via biparametric MRI radiomics. J Magnet Resonan Imaging JMRI. 2020;52:1102–9. Zhang L, Jiang D, Chen C, et al. Development and validation of a multiparametric MRI-based radiomics signature for distinguishing between indolent and aggressive prostate cancer. Br J Radiol. 2022;95:20210191. De Visschere P, Lumen N, Ost P, et al. Dynamic contrast-enhanced imaging has limited added value over T2-weighted imaging and diffusion-weighted imaging when using PI-RADSv2 for diagnosis of clinically significant prostate cancer in patients with elevated PSA. Clin Radiol. 2017;72:23–32. Fan X, **e N, Chen J, et al. Multiparametric MRI and machine learning based radiomic models for preoperative prediction of multiple biological characteristics in prostate cancer. Front Oncol. 2022;12: 839621. Qiao X, Gu X, Liu Y, et al. MRI radiomics-based machine learning models for Ki67 expression and gleason grade group prediction in prostate cancer. Cancers. 2023. https://doi.org/10.3390/cancers15184536. **g R, Wang J, Li J, et al. A wavelet features derived radiomics nomogram for prediction of malignant and benign early-stage lung nodules. Sci Rep. 2021;11:22330. Zheng Y, Zhou D, Liu H, et al. CT-based radiomics analysis of different machine learning models for differentiating benign and malignant parotid tumors. Eur Radiol. 2022;32:6953–64. Gu D, **e Y, Wei J, et al. MRI-based radiomics signature: a potential biomarker for identifying glypican 3-positive hepatocellular carcinoma. J Magnet Resonance Imaging JMRI. 2020;52:1679–87. Ogbonnaya CN, Zhang X, Alsaedi BSO, et al. Prediction of clinically significant cancer using radiomics features of pre-biopsy of multiparametric MRI in men suspected of prostate cancer. Cancers. 2021. https://doi.org/10.3390/cancers13246199. Lin Z, Wang T, Li Q, et al. Development and validation of MRI-based radiomics model to predict recurrence risk in patients with endometrial cancer: a multicenter study. Eur Radiol. 2023;33:5814–24. Xu L, Yang P, Liang W, et al. A radiomics approach based on support vector machine using MR images for preoperative lymph node status evaluation in intrahepatic cholangiocarcinoma. Theranostics. 2019;9:5374–85. Huang Y, Zhu T, Zhang X, et al. Longitudinal MRI-based fusion novel model predicts pathological complete response in breast cancer treated with neoadjuvant chemotherapy: a multicenter, retrospective study. EClinicalMedicine. 2023;58: 101899. Lefebvre TL, Ueno Y, Dohan A, et al. Development and validation of multiparametric MRI-based radiomics models for preoperative risk stratification of endometrial cancer. Radiology. 2022;305:375–86. Ma X, Gong J, Hu F, et al. Pretreatment multiparametric MRI-based radiomics analysis for the diagnosis of breast phyllodes tumors. J Magnet Resonan Imag JMRI. 2023;57:633–45. Tamada T, Kido A, Yamamoto A, et al. Comparison of biparametric and multiparametric MRI for clinically significant prostate cancer detection With PI-RADS version 2.1. J Magnetic Resonance Imaging JMRI. 2021;53:283–91. Russo F, Mazzetti S, Regge D, et al. Diagnostic accuracy of single-plane biparametric and multiparametric magnetic resonance imaging in prostate cancer: a randomized noninferiority trial in biopsy-naïve men. Eur Urol Oncol. 2021;4:855–62. Lee SS, Cheah YK. The interplay between microRNAs and cellular components of tumour microenvironment (TME) on non-small-cell lung cancer (NSCLC) progression. J Immunol Res. 2019;2019:3046379. Li H, Chen XL, Liu H, et al. MRI-based multiregional radiomics for preoperative prediction of tumor deposit and prognosis in resectable rectal cancer: a bicenter study. Eur Radiol. 2023. https://doi.org/10.1007/s00330-023-09723-9. Han C, Ma S, Liu X, et al. Radiomics models based on apparent diffusion coefficient maps for the prediction of high-grade prostate cancer at radical prostatectomy: comparison with preoperative biopsy. J Magnetic Resonan Imag : JMRI. 2021;54:1892–901. Zhou C, Zhang YF, Guo S, et al. Multiparametric MRI radiomics in prostate cancer for predicting Ki-67 expression and Gleason score: a multicenter retrospective study. Discov Oncol. 2023;14:133. The authors would like to thank all doctors, patients, and their family members for their support of the present study. This study was supported by the Bei**g Medical Award Foundation (YXJL-2022-0665-0197) and the grant from the Gansu Provincial Hospital (22GSSYD-33). Nini Pan: Project development, data collection, and manuscript writing. Liuyan Shi, Diliang He, Jianxin Zhao, Lianqiu ** Zhao: Project development. Gang Huang: Project development and manuscript editing. This study was approved by the ethics committee of the Gansu Provincial Hospital (2022–458) and Zhangye People's Hospital Affiliated to Hexi University, and all patients were exempted from signing an informed consent form. The authors declare that they have no competing interests. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional information, Intraclass correlation coefficient (ICC) results of all radiomics
features.
Additional Tables and Figures, Tables S1-S6, Figures S1-S3. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Pan, N., Shi, L., He, D. et al. Prediction of prostate cancer aggressiveness using magnetic resonance imaging radiomics: a dual-center study.
Discov Onc 15, 122 (2024). https://doi.org/10.1007/s12672-024-00980-8 Received: Accepted: Published: DOI: https://doi.org/10.1007/s12672-024-00980-8Data availability
References
Acknowledgements
Funding
Author information
Authors and Affiliations
Contributions
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Competing interests
Additional information
Publisher's Note
Supplementary Information
Rights and permissions
About this article
Cite this article
Keywords