Abstract
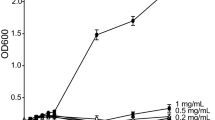
Bacillus species as fungal antagonistic agents have been widely used in the agriculture and considered as safe products for the management of plant pathogens. In this study, we reported the whole genome sequence of strain LJBV19 isolated from grapevine rhizosphere soil. Strain LJBV19 was identified as Bacillus velezensis through morphological, physicochemical, molecular analysis and genome comparison. Bacillus velezensis LJBV19 had a significant inhibitory effect on the growth of Magnaporthe oryzae with an inhibition ratio up to 75.55% and showed broad spectrum of activity against fungal phytopathogens. The 3,973,013-bp circular chromosome with an average GC content of 46.5% consisted of 3993 open reading frames (ORFs), and 3308 ORFs were classified into 19 cluster of orthologous groups of proteins (COG) categories. Genes related to cell wall degrading enzymes were predicted by Carbohydrate-Active enZYmes (CAZy) database and validated at the metabolic level, producing 0.53 ± 0.00 U/mL cellulose, 0.14 ± 0.01 U/mL chitinase, and 0.11 ± 0.01 U/mL chitosanase. Genome comparison confirmed the taxonomic position of LJBV19, conserved genomic structure, and genetic homogeneity. Moreover, 13 gene clusters for biosynthesis of secondary metabolites in LJBV19 genome were identified and two unique clusters (clusters 2 and 12) shown to direct an unknown compound were only present in strain LJBV19. In general, our results will provide insights into the antifungal mechanisms of Bacillus velezensis LJBV19 and further application of the strain.






Similar content being viewed by others
Availability of data and material
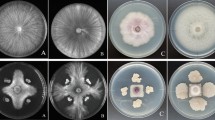
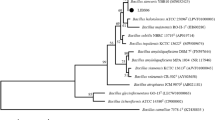
The GenBank accession number for the 16S rRNA gene of Bacillus velezensis LJBV19 is MZ157279. The complete sequence of the LJBV19 has been deposited in GenBank database under the accession number CP072563. The strain B. velezensis LJBV19 was publicly available from China General Microbiological Culture Collection Center (CGMCC) under the accession number CGMCC No. 21804. Morphological characteristics, phylogenetic tree, effect of LJBV19 on the mycelia of Botrytis cinerea, the location and products of secondary metabolite gene clusters, physiological and biochemical characteristics, minimum information about the genome sequence, genome-to-genome comparison (ANI and dDDH), and inhibition radius of B. velezensis LJBV19 against 12 pathogens are available as Supplementary Materials.
References
Adeniji AA, Loots DT, Babalola OO (2019) Bacillus velezensis: phylogeny, useful applications, and avenues for exploitation. Appl Microbiol Biotechnol 103(9):3669–3682. https://doi.org/10.1007/s00253-019-09710-5
Anckaert A, Arias AA, Hoff G, Calonne-Salmon M, Declerck S, Ongena M (2021) The use of Bacillus spp. as bacterial biocontrol agents to control plant diseases. Microbial bioprotectants for plant disease management. Burleigh Dodds Series in Agricultural Science. Burleigh Dodds Science Publishing Limited, pp 247–300. https://doi.org/10.19103/as.2021.0093.10
Andrić S, Meyer T, Ongena M (2020) Bacillus responses to plant-associated fungal and bacterial communities. Front Microbiol 11:1350. https://doi.org/10.3389/fmicb.2020.0135
Garrity G (2001) Bergey's manual of systematic bacteriology 38(4):443–491
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene Ontology C (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340(4):783–795. https://doi.org/10.1016/j.jmb.2004.05.028
Berg G (2009) Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84(1):11–18. https://doi.org/10.1007/s00253-009-2092-7
Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FSL, Simon Fraser Univ Res Comp G (2017) IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45(W1):W30–W35. https://doi.org/10.1093/nar/gkx343
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29(12):2607–2618. https://doi.org/10.1093/nar/29.12.2607
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T (2021) antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49(W1):W29–W35. https://doi.org/10.1093/nar/gkab335
Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31(1):365–370. https://doi.org/10.1093/nar/gkg095
Borriss R, Chen X-H, Rueckert C, Blom J, Becker A, Baumgarth B, Fan B, Pukall R, Schumann P, Sproeer C, Junge H, Vater J, Puehler A, Klenk H-P (2011) Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7(T) and FZB42(T): a proposal for Bacillus amyloliquefaciens subsp amyloliquefaciens subsp nov and Bacillus amyloliquefaciens subsp plantarum subsp nov based on complete genome sequence comparisons. Int J Syst Evol Microbiol 61:1786–1801. https://doi.org/10.1099/ijs.0.023267-0
Borriss R, Danchin A, Harwood CR, Medigue C, Rocha EPC, Sekowska A, Vallenet D (2018) Bacillus subtilis, the model Gram-positive bacterium: 20 years of annotation refinement. Microb Biotechnol 11(1):3–17. https://doi.org/10.1111/1751-7915.13043
Cai XC, Liu CH, Wang BT, Xue YR (2017) Genomic and metabolic traits endow Bacillus velezensis CC09 with a potential biocontrol agent in control of wheat powdery mildew disease. Microbiol Res 196:89–94. https://doi.org/10.1016/j.micres.2016.12.007
Chen K, Tian Z, He H, Long C-A, Jiang F (2020) Bacillus species as potential biocontrol agents against citrus diseases. Biol Control. https://doi.org/10.1016/j.biocontrol.2020.104419
Chen W-C, Juang R-S, Wei Y-H (2015) Applications of a lipopeptide biosurfactant, surfactin, produced by microorganisms. Biochem Eng J 103:158–169. https://doi.org/10.1016/j.bej.2015.07.009
Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Suessmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R (2007) Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25(9):1007–1014. https://doi.org/10.1038/nbt1325
Chen YJ, Yu P, Luo JC, Jiang Y (2003) Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm Genome 14(12):859–865. https://doi.org/10.1007/s00335-003-2296-6
Chen ZY, Abuduaini X, Mamat N, Yang QL, Wu MJ, Lin XR, Wang R, Lin RR, Zeng WJ, Ning HC, Zhao HP, Li JY, Zhao HX (2021) Genome sequencing and functional annotation of Bacillus sp. strain BS-Z15 isolated from cotton rhizosphere soil having antagonistic activity against Verticillium dahliae. Arch Microbiol 203(4):1565–1575. https://doi.org/10.1007/s00203-020-02149-7
Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, Cramer GR, Delledonne M, Luo C, Ecker JR, Cantu D, Rank DR, Schatz MC (2016) Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13(12):1050–1054. https://doi.org/10.1038/nmeth.4035
Choo KH, Tong JC, Zhang L (2004) Recent applications of hidden Markov models in computational biology. Genomics Proteomics Bioinformatics 2(2):84–96
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68(1):461–466. https://doi.org/10.1099/ijsem.0.002516
Compant S, Duffy B, Nowak J, Clement C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71(9):4951–4959. https://doi.org/10.1128/aem.71.9.4951-4959.2005
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5(6):e11147. https://doi.org/10.1371/journal.pone.0011147
Dhouib H, Zouari I, Ben Abdallah D, Belbahri L, Taktak W, Triki MA, Tounsi S (2019) Potential of a novel endophytic Bacillus velezensis in tomato growth promotion and protection against Verticillium wilt disease. Biol Control 139:11. https://doi.org/10.1016/j.biocontrol.2019.104092
Dufour A, Hindre T, Haras D, Le Pennec JP (2007) The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol Rev 31(2):134–167. https://doi.org/10.1111/j.1574-6976.2006.00045.x
Dunlap CA, Kim SJ, Kwon SW, Rooney AP (2016) Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int J Syst Evol Microbiol 66(3):1212–1217. https://doi.org/10.1099/ijsem.0.000858
Eyles TH, Vior NM, Lacret R, Truman AW (2021) Understanding thioamitide biosynthesis using pathway engineering and untargeted metabolomics. Chem Sci 12(20):7138–7150. https://doi.org/10.1039/d0sc06835g
Fan B, Blom J, Klenk H-P, Borriss R (2017) Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an “Operational Group B. amyloliquefaciens” within the B-subtilis species complex. Front Microbiol 8:22. https://doi.org/10.3389/fmicb.2017.00022
Fan B, Wang C, Song X, Ding X, Wu L, Wu H, Gao X, Borriss R (2018) Bacillus velezensis FZB42 in 2018: The Gram-positive model strain for plant growth promotion and biocontrol. Front Microbiol 9:2491. https://doi.org/10.3389/fmicb.2018.02491
Gao Z, Zhang B, Liu H, Han J, Zhang Y (2017) Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol Control 105:27–39. https://doi.org/10.1016/j.biocontrol.2016.11.007
Grissa I, Vergnaud G, Pourcel C (2008) CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 36:W145–W148. https://doi.org/10.1093/nar/gkn228
Hamaoka K, Aoki Y, Suzuki S (2021) Isolation and characterization of endophyte Bacillus velezensis KOF112 from grapevine shoot xylem as biological control agent for fungal diseases. Plants 10(9):1815. https://doi.org/10.3390/plants10091815
Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P (2008) eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res 36:D250–D254. https://doi.org/10.1093/nar/gkm796
Jiang C-H, Liao M-J, Wang H-K, Zheng M-Z, Xu J-J, Guo J-H (2018) Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol Control 126:147–157. https://doi.org/10.1016/j.biocontrol.2018.07.017
Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI (2018) Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics 62(1):e51–e51. https://doi.org/10.1002/cpbi.51
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44(D1):D457–D462. https://doi.org/10.1093/nar/gkv1070
Keswani C, Singh HB, Garcia-Estrada C, Caradus J, He Y-W, Mezaache-Aichour S, Glare TR, Borriss R, Sansinenea E (2020) Antimicrobial secondary metabolites from agriculturally important bacteria as next-generation pesticides. Appl Microbiol Biotechnol 104(3):1013–1034. https://doi.org/10.1007/s00253-019-10300-8
Khan A, Singh P, Srivastava A (2018) Synthesis, nature and utility of universal iron chelator - siderophore: a review. Microbiol Res 212–213:103–111. https://doi.org/10.1016/j.micres.2017.10.012
Kim DR, Jeon CW, Shin JH, Weller DM, Thomashow L, Kwak YS (2019) Function and distribution of a lantipeptide in strawberry Fusarium wilt disease-suppressive soils. Mol Plant Microbe Interact 32(3):306–312. https://doi.org/10.1094/mpmi-05-18-0129-r
Kudo F, Numakura M, Tamegai H, Yamamoto H, Eguchi T, Kakinuma K (2005) Extended sequence and functional analysis of the butirosin biosynthetic gene cluster in Bacillus circulans SANK 72073. J Antibiot 58(6):373–379. https://doi.org/10.1038/ja.2005.47
Lagesen K, Hallin P, Rodland EA, Staerfeldt H-H, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35(9):3100–3108. https://doi.org/10.1093/nar/gkm160
Li WZ, Jaroszewski L, Godzik A (2002) Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18(1):77–82. https://doi.org/10.1093/bioinformatics/18.1.77
Li Y, Heloir MC, Zhang X, Geissler M, Trouvelot S, Jacquens L, Henkel M, Su X, Fang XW, Wang Q, Adrian M (2019) Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol Plant Pathol 20(8):1037–1050. https://doi.org/10.1111/mpp.12809
Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42(D1):D490–D495. https://doi.org/10.1093/nar/gkt1178
Lopes R, Tsui S, Goncalves PJRO, de Queiroz MV (2018) A look into a multifunctional toolbox: endophytic Bacillus species provide broad and underexploited benefits for plants. World J Microbiol Biotechnol 34(7):94. https://doi.org/10.1007/s11274-018-2479-7
Lowe TM, Eddy SR (1997) tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964. https://doi.org/10.1093/nar/25.5.955
Moldenhauer J, Goetz DCG, Albert CR, Bischof SK, Schneider K, Suessmuth RD, Engeser M, Gross H, Bringmann G, Piel J (2010) The final steps of Bacillaene biosynthesis in Bacillus amyloliquefaciens FZB42: direct evidence for beta, gamma dehydration by a trans-acyltransferase polyketide synthase. Angew Chem Int Ed 49(8):1465–1467. https://doi.org/10.1002/anie.200905468
Mullins AJ, Li Y, Qin L, Hu X, **e L, Gu C, Mahenthiralingam E, Liao X, Webster G (2020) Reclassification of the biocontrol agents Bacillus subtilis BY-2 and Tu-100 as Bacillus velezensis and insights into the genomic and specialized metabolite diversity of the species. Microbiology 166(12):1121–1128. https://doi.org/10.1099/mic.0.000986
Nannan C, Vu HQ, Gillis A, Caulier S, Nguyen TTT, Mahillon J (2021) Bacilysin within the Bacillus subtilis group: gene prevalence versus antagonistic activity against Gram-negative foodborne pathogens. J Biotechnol 327:28–35. https://doi.org/10.1016/j.jbiotec.2020.12.017
Olishevska S, Nickzad A, Deziel E (2019) Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl Microbiol Biotechnol 103(3):1189–1215. https://doi.org/10.1007/s00253-018-9541-0
Ongena M, Jacques P (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16(3):115–125. https://doi.org/10.1016/j.tim.2007.12.009
Ortega MA, van der Donk WA (2016) New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem Biol 23(1):31–44. https://doi.org/10.1016/j.chembiol.2015.11.012
Palazzini JM, Dunlap CA, Bowman MJ, Chulze SN (2016) Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol Res 192:30–36. https://doi.org/10.1016/j.micres.2016.06.002
Ravi S, Nakkeeran S, Saranya N, Senthilraja C, Renukadevi P, Krishnamoorthy AS, El Enshasy HA, El-Adawi H, Malathi VG, Salmen SH, Ansari MJ, Khan N, Sayyed RZ (2021) Mining the genome of Bacillus velezensis VB7 (CP047587) for MAMP genes and non-ribosomal peptide synthetase gene clusters conferring antiviral and antifungal activity. Microorganisms 9(12):2511. https://doi.org/10.3390/microorganisms9122511
Rabbee MF, Ali MS, Choi J, Hwang BS, Jeong SC, Baek KH (2019) Bacillus velezensis: a valuable member of bioactive molecules within plant microbiomes. Molecules 24(6):1046. https://doi.org/10.3390/molecules24061046
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106(45):19126–19131. https://doi.org/10.1073/pnas.0906412106
Rooney AP, Price NP, Ehrhardt C, Swezey JL, Bannan JD (2009) Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol 59(Pt 10):2429–2436. https://doi.org/10.1099/ijs.0.009126-0
Santana MA, Moccia-V CC, Gillis AE (2008) Bacillus thuringiensis improved isolation methodology from soil samples. J Microbiol Methods 75(2):357–358. https://doi.org/10.1016/j.mimet.2008.06.008
Sari GL, Trihadiningrum Y, Ni’matuzahroh (2019) Isolation and identification of native bacteria from total petroleum hydrocarbon polluted soil in Wonocolo public oilfields, Indonesia. J Ecol Eng 20(8):60–64. https://doi.org/10.12911/22998993/110816
Sasse J, Martinoia E, Northen T (2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23(1):25–41. https://doi.org/10.1016/j.tplants.2017.09.003
Shafi J, Tian H, Ji M (2017) Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotechnol Equip 31(3):446–459. https://doi.org/10.1080/13102818.2017.1286950
Ton That Huu D, Nguyen Thi Kim C, Pham Viet C, Smidt H, Sipkema D (2021) Diversity and antimicrobial activity of Vietnamese sponge-associated bacteria. Marine Drugs 19(7):353. https://doi.org/10.3390/md19070353
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. Plos One 9(11):e112963. https://doi.org/10.1371/journal.pone.0112963
Wang B, Peng H, Wu W, Yang B, Chen Y, Xu F, Peng Y, Qin Y, Lu J, Fu P (2021) Genomic insights into biocontrol potential of Bacillus stercoris LJBS06. 3 Biotech 11:458. https://doi.org/10.1007/s13205-021-03000-6
Ye M, Tang X, Yang R, Zhang H, Li F, Tao F, Li F, Wang Z (2018) Characteristics and application of a novel species of Bacillus: Bacillus velezensis. ACS Chem Biol 13(3):500–505. https://doi.org/10.1021/acschembio.7b00874
Zhang M, Li J, Shen A, Tan S, Yan Z, Yu Y, Xue Z, Tan T, Zeng L (2016) Isolation and Identification of Bacillus amyloliquefaciens IBFCBF-1 with potential for biological control of Phytophthora blight and growth promotion of pepper. J Phytopathol 164(11–12):1012–1021. https://doi.org/10.1111/jph.12522
Zhu XF, Zhou Y, Feng JL (2007) Analysis of both chitinase and chitosanase produced by Sphingomonas sp. CJ-5. J Zhejiang Univ Sci B 8(11):831–838. https://doi.org/10.1631/jzus.2007.B0831
Funding
This work was supported by Shanghai Municipal Agricultural Commission (grant no. 2021–02-08–00-12-F00751), National Key R & D Program of China (2019YFD1002501 and 2018YFD1000301), Shanghai Municipal Commission for Science and Technology (18391900400), Yunnan Province Science and Technology Department (202005AF150023 to J. Lu), and China Agriculture Research System (grant no. CARS-29-yc-2).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Bo Wang, Bohan Yang, Hang Peng, and Peining Fu. The first draft of the manuscript was written by Bo Wang and Bohan Yang; all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
All authors approved the manuscript.
Consent for publication
Written informed consent for publication was obtained from all participants.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bo Wang and Bohan Yang contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, B., Yang, B., Peng, H. et al. Genome sequence and comparative analysis of fungal antagonistic strain Bacillus velezensis LJBV19. Folia Microbiol 68, 73–86 (2023). https://doi.org/10.1007/s12223-022-00996-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-022-00996-z