Abstract
Biodiversity related observational data are collected in a variety of ways and for a variety of purposes, mostly in the form of some sort of organised data collection action. Data management solutions are often developed to manage the data collection processes and organise the data, which may work well on their own but are less compatible with other data management tools.
In a continuous development process, we have created the OpenBioMaps (OBM) biodiversity data management platform, which can be used as a self-hosted data management platform and as a free service, hosted by several institutions for biological database projects. OBM has the ability to integrate biological databases without any structural or functional constraints, allowing a high degree of flexibility in data management and development; it provides interfaces to facilitate communication between different end-user communities, including scientists, citizens, conservationists and educational staff. We have also established a network of OBM services based on collaboration between government, educational and scientific institutions and NGOs to provide a public service to those who lack the capacity or knowledge to set up or manage their own self-hosted servers.
OpenBioMaps uniquely focuses on the entire data management process, from building the data structure to data collection, visualisation, sharing and processing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Management of biodiversity data varies considerably depending on its purposes, tools and user community. Although there is a broad consensus that data should be made the most accessible for researchers (e.g. Nelson 2009; Costello et al. 2014; Powers and Hampton 2019) and there are a number of recommendations (Moritz et al. 2011; Costello et al. 2014; Costello and Wieczorek 2014) and tools (e.g. Mudunuri et al. 2009; Wieczorek et al. 2012; Robertson et al. 2014; Global Biodiversity Information Facility, GBIF 2016; Clark et al. 2016) in this regard, however, there are no general solutions for managing data, especially in everyday work environments, that can greatly facilitate this.
Daily work with biodiversity data usually involves the management of incoming data regardless of the way, size, or purpose of the data collection. An essential parameter of biodiversity data is their spatial location, so one of the most common management requirements is to display the species occurances on a map. In addition, the ability to sort, query, and authorise data access are highlighted as general and common requirements for these systems.
There are two important focus groups in terms of usage: the data collectors and the data curators (Tammaro et al. 2019). In a well-functioning data management system, the needs of both collectors and curators must be considered during development, otherwise the data management process will become hampered and hence slow.
In addition to meeting all these conditions, other indispensable goals may emerge, especially for long-term data collection projects, such as linking data or certain workflows to other tools or processes and thus ensuring integration (Heberling et al. 2021).
Thanks to the large number of open source and freely available commercial or community-based tools and solutions, it is quite easy to build a new workflow for biodiversity data management that meets the above criteria. However, maintaining a private system in the long run is usually not a simple task (Bastow and Leonelli 2010; Reichman et al. 2011). Nevertheless, it is common practice to develop a new, private data management workflow. For example, in Hungary, where millions of new biodiversity monitoring related occurrence data are recorded annually by employees of the regional nature conservation authorities, the national parks direcorates, many different data management systems existed side by side. Other Hungarian governmental organisations such as the Ministry of Agriculture also developed their own biodiversity data management system (Takács and Szilágyi 2004). In addition, Hungarian conservation NGOs also have developed private management solutions, the details of which depend on their financial or technical background. Furthermore, researchers from scientific institutes and university research groups are also develo** custom tools to implement data management processes (e.g. Molnár etal 2007). Generally speaking, the technical diversity of data management practices discourages the development of collaborations, which would, however, be very desirable (e.g. McShane et al. 2004, Bennett et al. 2017, Carmen et al. 2015). Therefore, new collaborations usually require the development or creation of new tools and interfaces for data exchange or sharing, which hinders the implementation of the FAIR (Findability, Accessibility, Interoperability, Reusability) principles (Wilkinson et al. 2016), and resulting leaving large amounts of data in the ‘dark’ (Heidorn 2008) in more or less isolated databases. In most cases these isolated databases are hard to discover and are inaccessible to the public (Blair et al. 2020), sometimes even to the target group whom it is created for.
Our aim was to develop a complex and flexible software solution that supports the management of biodiversity data throughout its lifecycle and is suitable for handling both public and sensitive data, further facilitating communication between conservationists, educators, government officials and scientists. We also created a public and free service based on this software components for building easily manageable biodiversity-related databases, which then can facilitate the open sharing of data. All these software components are compatible with open standards (e.g. Wieczorek et al. 2012) and entirely based on open-source solutions and customizable with user-contributed extensions. Finally, we have initiated and developed a formal collaboration that encompasses several institutes (following the ‘bigger is better’ advice (Costello et al. 2014)) to guarantee the long-term support of these services and to oversee the software development.
Design and implementation
Rationale for the design of the system architecture
OpenBioMaps is a grassroots project and does not have a fixed budget. All features and tools are driven by user needs. Economic sustainability was therefore a key factor in the design of the OpenBioMaps system. In addition, we were designing for a user community whose members (researchers and conservationists) already had tools and routines to perform certain tasks, to which our newly created tools had to adapt. Therefore, all the tools we developed are characterised by a low level of connectivity support and a high level of flexibility.
For the above reasons, we have developed using only open source and freely available tools and do not rely on any large development framework to keep external dependencies low. Although we follow mainstream development practices, we always choose the most stable and maintainable solutions.
The main server components (such as PostgreSQL and MapServer) are externally accessible and can be integrated into independent systems. In addition to this support for technical accessibility, the creation of SQL structures supports interoperability by creating structures for data managed in OpenBioMaps that are understandable to humans and can be used in other application environments without any special transformations. In other words, the structure of the managed data is only minimally complemented by OpenBioMaps and is basically optimised for interoperable data entry and querying.
Application logic and tools
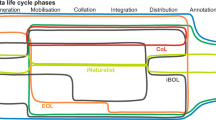
OpenBioMaps is mostly used to digitize and manage biodiversity data collected in the field. Its toolkit follows the life cycle of the data and offers a range of customisable solutions at all levels (Fig. 1). These levels are: (1) database initialisation, (2) filling the database with data, (3) data curation and organisation, (4) data processing and evaluation and (5) data sharing and supporting the publication process.
(1) Database projects with new custom structure and settings (access, visualisation, upload-forms) can be created via the Web UI. And can be maintained through a Web based system administrator application and there is and other Web UI application for the project management. (2) Data collection is supported by a mobile application that allows offline data collection for Android and iPhone users. The mobile application manages data collection forms according to the settings of individual database projects and can display the collected data. Data collected by other tools can also be uploaded through the Web UI, also using customisable forms that allows for the processing of various files. (3) Once uploaded, the data can be further organised and transformed (e.g. validation, extending attributes, adding metadata) automatically using background processes (any kind of non-interactive software can be integrated as a background job), or manully using map or data filters and modification tools. (4) Data stored in the database can be accessed from Postgres/Postgis client applications and the map data can be accessed by WMS, WFS clients. All the stored data can be accessed with external tools through OBM API which let users integrate the data workflow into external applications. There is an OBM R package that makes data access easy and scriptable in an R environment, and a built-in R Shiny application interface that helps to review the contents of data tables using interactive charts. Also, there is an interface to run analysis remotely, managing access to distributed computing capacity and providing tool support for computationally intensive analyses. (5) Finally, Web UI provides interface to place analyses files, processed data or raw data in remote repositories with detailed metadata to support publication tasks. Web UI also provides interface with persistent identifiers for raw data sharing. These features help improve the data findability and, while also, opens the way towards the reusability.
System architecture
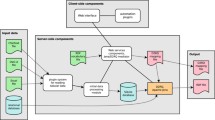
The architecture of the system is presented from the perspective of the central web application (Fig. 2). This is a system of more or less independent web applications with multiple entry points, forming a unified system with connections built across an internal API and an external API. These independent applications are (Table 1):
(1) Map and text query and data management interface (Fig. 3). The map interface is built using MapServer through an internal MapProxy application, which is an authorization tool in front of Mapserver; (2) Data upload interface (file uploads and web forms); (3) Project administration interface (user profile settings, invitations, new project creation, module settings and all additional project settings); (4) System administration application where system updates and maintenance can be performed.
The External API allows data queries and data uploads to external applications. The Internal API is an API interface, called Rest API but without web requests, for managing standardised communication between separate internal components. The web application suite can be installed using Docker environment for great portability and flexibility and this lets users easily create a self-hosted database portal.
The overall system also includes a number of other tools, which are best characterised by their place in the data processing workflow, as they are not mandatory elements of the core system. These components can be connected to the core system elements through internal or external APIs and their presence depends on the specific design of the project (Table 2).
These applications are:
Mobile application for fieldwork by biologists (Fig. 4), R-package, R-Shiny application and compute server application for data analysis.
Modules to extend the functionality of the web user interfaces. Some modules allow the integration of additional custom software components (plug-ins), while all modules can be replaced by custom code versions on demand. There are currently more than 40 modules.
The remote repository interface (currently implemented only for the Dataverse system) communicates with the remote repository API via an internal repository proxy, over which it creates its own management layer. This is necessary to allow a project to communicate with the repository system through a single repository user, instead of registering individual users.
The GIS Map** web application is a tool open for external use and is involved in several internal processes. This application has its own API.
Finally, Jobs are standalone applications, controlled by a scheduler and run in the background. Jobs have their own GIT repository, which is directly accessible from projects, thus supporting dynamic custom development. The retrieved Jobs are often cutomised with the integrated CodeMirror based text editor.
Public online service
Although OBM is a portable system and many institutions deployed private OBM servers to serve their own projects, some large institutes have set up public servers to provide freely available capacity for smaller projects. Currently, there are four OBM public server nodes, hosted by two universities (University of Debrecen and Eötvös Lóránd University), the Duna-Ipoly National Park Directorate and the European Open Science Cloud (EOSC). These OBM nodes are interconnected with each other via a private network forming the OBM network to share some resources, e.g. backup storage or computational capacity.
A list of known servers is available in an OpenBioMaps database created for this purpose. Servers registered in the open OpenBioMaps Networks Database (2016) can be accessed from the mobile application.
Examples of usage and facilities
There are currently about 60 active database projects running on 11 independent OBM servers, which approximately contain seven million records with spatial reference of about 20,000 species from various biodiversity data collecting studies. A list of known servers and databases is available from openbiomaps.org. In this list, databases are categorised as stable, test or experimental, which is only indicative information about the maturity of the database. An additional flag is used to indicate the openness of the database and the data it contains. This flag indicates how open the database is to uploading, reading and modification.
The largest users of OBM are currently the Hungarian national park directorates and nature conservation NGOs. In addition OBM is used in research projects e.g. the “Sex Ratio in Vertebrates” database (Liker et al. 2021, Sex Ratio in Vertebrates database 2017); Global Plover Database (Halimubieke et al. 2020; Global Plover Database 2017), Debrecen DNA Bank (2018), Flora Atlas of Hungary (2018) and Balkanherps (2017). There are also several examples of using OBM in CitizenScience projects, e.g. Eurasian Kurgan Database (Deák et al. 2019, Eurasian Kurgan Database 2019), OpenBirdMaps (2013) and OpenHerpMaps (2019) in Romania and Public Nestbox Breeding Data (2016) in Hungary. Finally, there are examples of use in educational projects e.g. Birds of Debrecen (2019) where student collaborative work and field data collection can be performed on OBM basis.
Examples of workflows using OpenBioMaps
One of the largest OBM project currently is the Pannon Bioregion Biotic Dataset (2012), assembled by the Danube-Ipoly National Park Directorate in Hungary. This spatial database contains more than two million occurrence records of about 10 000 different species. Majority of the records were collected in the field using handheld GPS devices, with high spatial accuracy. However, it also comprises records of natural history collections with weak spatial accuracy, some of which are over 100 years old. Recently, most of the data has been collected through the OBM mobile application, by both professionals and amateurs (volunteer data collectors). OBM instances operated by other national parks directorates, replaced a previous data management system and integrated all data from there. A significant amount of records has been uploaded from data files produced in other applications.
Data curators play a key role in the system, as they do the bulk of the non-mobile data uploads, review the data input and liaise with data collectors. In addition, they create a number of data collection forms for each data collection and, finally, they continuously organise and populate the SQL database with data from other sources (e.g. raster maps, official government databases), which data users use mostly in QGIS or OpenBioMaps environments to carry out their daily work. This database is closed to the public, but data for certain questions is shared upon request.
The largest public database is OpenBirdMaps (2013), operated by the Milvus Group Association, an NGO in Romania. It is dedicated to collect and freely publish information regarding the spatial and temporal distribution of wild birds in Romania. In April 2022 about 80% of the total number of 1,511,000 records from this database were public (excluding sensitive data). It has over 300 users, who mostly upload their records through the OBM mobile application. The curators have built a number of data management tools that can be used to perform automated daily routine tasks around the database, such as simple checks and corrections or linking observation-list data and transferring the data to other databases in a specific format. OpenBirdMaps is a partner of the EuroBirdPortal initiative, a project run by the European Bird Census Council (EBCC 2022) in partnership with more than 80 institutions from 29 countries in Europe. Data transfer to EuroBirdPortal is an automated daily process using a background job.
In the Hunviphab (2021) project, a LIFE project (https://www.rakosivipera.hu/), monitoring coordinators are carrying out at least seven different monitoring programmes on around 5,000 hectares, involving more than 30 volunteers and professionals each year, from February to November. Data is collected using the OBM mobile app and DCLR cameras. Data collected with mobile phones are usually automatically uploaded to the database (depending on the setting) and participants check and update their field data as needed (e.g. uploading and matching camera photos with data). In this project, tracklogs of data collectors in the field are critical information for calculating accurate search effort, so the tracklogging feature of the mobile app is used to collect large amounts of tracklog data. The monitoring coordinators, as curators of the data, have created several R-scripts available in the hunviphab R package to continuously process and validate the different types of data that are coming in. Their R-scripts use the OBM R package to retrieve data from databases, and use a number of database-level functions to transform the incoming data (e.g. compiling tracklogs with observations), and preprocess datasets for statistical analyses such as abundance estimation applying n-mixture models for the focal species, the Hungarian meadow viper (Vipera ursinii rakosiensis).
The Eurasian Kurgan Database (Deák et al. 2019, Eurasian Kurgan Database 2019) has two types of data, detailed and simple. They are managed in the same data table, but have different map views and can be uploaded using two separate forms available as mobile field and web-based file upload forms. The simple data form is publicly available and contains only the coordinate, date, observer and optionally a photo field, while the detailed data form is only available to project members and contains the parameters for a detailed survey of a kurgan (botanical and environmental condition survey). In this way, the database is partially open for uploading and reading, as the simplified version of the data is freely accessible, while the detailed data is only available to members.
The majority of the data is received from Asian and European countries in excel files, which are uploaded by the data curators using the file upload version of the forms above.
The data has been checked once so far, and this process took place outside OpenBioMaps, so the entire database was re-imported at the end of the validation process.
Anyone can register their nest box in the Public NestBox Breeding Data (2016) and upload their breeding observation data. However, this database is currently mostly used for research projects around the University of Debrecen campus, where there are about 400 wooden nesting boxes from various research and citizen projects over the last 20 years. These nest boxes are located in an area of about 50 hectares around the campus and are difficult to monitor during the breeding season.
The project curators created online colour maps (using Mapserver in the OBM interface) and automatically updated online tables based on database queries for fieldwork students to visualise and organise the following monitoring events, to ensure that all observations are made in a timely manner, as even a day’s delay can mean a complete data set is lost. In addition to the custom forms created in the mobile apps, fieldwork students also use the online database interface to keep track of the most up-to-date information available on nest boxes.
They use different data collection methods depending on the type of experiments and lab work, and create custom data tables for specific examinations, but link it all to the main tables of the projects, basically PostgreSQL level solutions.
The data structure and SQL solutions developed in this project have been adopted in several other research projects using OBM, where regular observations are linked to permanent observation sites and these observation events can be accompanied by additional associated event data tables.
Discussion
We have developed an online, non-centralised system based on open source tools to provide a sustainable and reliable framework for the various communities that develop and operate biodiversity-related databases. The OBM provides flexible interfaces to standard back-end services (relational database, map server) and includes a number of tools to facilitate the development of biodiversity data structures and the exchange of information stored in the databases with other applications and databases. We have also created a reliable public service based on this system for those who would like to run self maintained data management projects without their own infrastructure. These infrastructures and services can do much to promote the use of biodiversity data, in particular by significantly accelerating the data lifecycle from generation to use, while also contributing to the implementation of the FAIR principles.
Software availability and requirements
The OpenBioMaps server can be self hosted, or can be deployed on cloud architecture using the same Docker based installation method. The software components of the OpenBioMaps platform for a self hosted deployment are available through GitLab (https://gitlab.com/openbiomaps) and GitHub (https://github.com/openbiomaps). This repositories are group repositories which includes the Docker description of the server installation (docker/obm-composer), web application (obm web-app),web application modules (web-app modules), job applications (web-app jobs), customisable landing page application (obm-web-mainpages), and customisable style application for web ui (openbiomaps-styles). Furthermore, these repositories are hosting OpenBioMaps client applications such as the Mobile application (openbiomaps-mobile), R package (obm.r), R Shiny application (obm-rshiny) and the Sphynx based documentation (documentation).
To maintain a self-hosted private OBM server in a production environment, we recommend a minimum of 4 CPU cores, at least 8 Gb of memory and 100 Gb of storage space for smooth operation.
The internationalisation of the entire platform is publicly available through a private Weblate instance (a community-based translation application) on https://translations.openbiomaps.org.
The cloud based deployment of OpenBioMaps server is available via EOSC Marketplace (https://marketplace.eosc-portal.eu/services/openbiomaps), where different deploying options offered from the automatic server deploying on cloud infrastructure to different kind of server intergrations.
The OBM mobile application is available through Google Play and Apple’s App Store.
References
Balkanherps (2017) – Available at https://openbiomaps.org/projects/balkanherps/
Bastow R, Leonelli S (2010) Sustainable digital infrastructure. Although databases and other online resources have become a central tool for biological research, their long-term support and maintenance is far from secure. EMBO Rep 11:730–734. https://doi.org/10.1038/embor.2010.145
Bennett NJ, Roth R, Klain SC, Chan KMA, Clark DA, Cullman G, Epstein G, Nelson MP, Stedman R, Teel TL, Thomas REW, Wyborn C, Curran D, Greenberg A, Sandlos J, Veríssimo D (2017) Mainstreaming the social sciences in conservation. Conserv Biol 31:56–66
Birds of Debrecen (2019) – Available at https://openbiomaps.org/projects/debrecenmadarai/
Blair J, Gwiazdowski R, Borrelli A, Hotchkiss M, Park C, Perrett G, Hanner R (2020) Towards a catalogue of biodiversity databases: An ontological case study. Biodivers Data J 8:e32765. https://doi.org/10.3897/BDJ.8.e32765
Carmen E, Nesshöver C, Saarikoski H, Vandewalle M, Watt A, Wittmer H, Young J (2015) Creating a biodiversity science community: experiences from a European Network of Knowledge. Environ Sci Policy 54:497–504
Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2016) GenBank Nucleic Acids Res 44:D67–D72. https://doi.org/10.1093/nar/gkv1276
Costello MJ, Appeltans W, Bailly N, Berendsohn WG, de Jong Y, Edwards M, Froese R, Huettmann F, Los W, Mees J, Segers H, Bisby FA (2014) Strategies for the sustainability of online open-access biodiversity databases. Biol Conserv 173:155–165
Costello MJ, Wieczorek J (2014) Best practice for biodiversity data management and publication. Biol Conserv 173:68–73
Deák B, Tóth CA, Bede Á, Apostolova I, Bragina TM, Báthori F, Bán M (2019) Eurasian Kurgan Database – a citizen science tool for conserving grasslands on historical sites. Hacquetia 18(2):179–187
Debrecen DNA Bank (2018) – Available at https://openbiomaps.org/projects/debrecen_dnabank/
EBCC - European Bird Census Council (2022) EuroBirdPortal. Available at https://eurobirdportal.org (accessed April 2022)
Eurasian Kurgan Database (2019) – Available at https://openbiomaps.org/projects/kurgan/, https://doi.org/10.18426/obm.3mbbectm2bmg
Flora Atlas of Hungary (2018) – Available at http://floraatlasz.uni-sopron.hu, https://doi.org/10.18426/obm.17ga3jhgu710
GBIF (2016) GBIF portal. Available at http://www.gbif.org (accessed April 2022)
Global Plover Database (2017) – Available at https://openbiomaps.org/projects/plover/
Halimubieke N, Kupán K, Valdebenito JO et al (2020) Successful breeding predicts divorce in plovers. Sci Rep 10:15576. https://doi.org/10.1038/s41598-020-72521-6
Heidorn PB (2008) Shedding light on the dark data in the long tail of science. Libr Trends 57(2):280–299
Heberling JM, Miller JT, Noesgaard D, Weingart SB, Schigel D (2021) Data integration enables global biodiversity synthesis. PNAS 118. https://doi.org/10.1073/pnas.2018093118
HUNVIPHAB (2021) – Available at https://knp.openbiomaps.org/projects/hunviphab/
Liker A, Bókony V, Pipoly I, Lemaître JF, Gaillard JM, Székely T, Freckleton RP (2021) Evolution of large males is associated with female-skewed adult sex ratios in amniotes. Evolution 75:1636–1649. https://doi.org/10.1111/evo.14273
MapServer v7.2.3 (2021) Open Source geographic data rendering engine. Home page, http://mapserver.gis.umn.edu/. Accessed at April 2021
McShane TO, Wells MP (2004) Getting biodiversity projects to work: towards more effective conservation and development. Columbia University Press
Molnár Z, Bartha S, Seregélyes T, Illyés E, Botta-Dukát Z, Tímár G, Horváth F, Révész A, Kun A, Bölöni J, Biró M, Bodonczi L, Deák ÁJ, Fogarasi P, Horváth A, Isépy I, Karas L, Kecskés F, Molnár C, Rév Sz (2007) A grid-based, satellite-image supported multi-attributed vegetation map** method (MÉTA). Folia Geobotanica 42:225–247
Moritz T, Krishnan S, Roberts D, Ingwersen P, Agosti D, Penev L, Cockerill M, Chavan V (2011) Towards mainstreaming of biodiversity data publishing: recommendations of the GBIF Data Publishing Framework Task Group. BMC Bioinformatics 12:S1. https://doi.org/10.1186/1471-2105-12-S15-S1
Mudunuri U, Che A, Yi M, Stephens RM (2009) bioDBnet: the biological database network. Bioinformatics 25(4):555–556
Nelson B (2009) Data sharing: Empty archives. Nature 461:160–163. https://doi.org/10.1038/461160a
OpenBioMaps Network Database (2016) – Available at https://openbiomaps.org/projects/openbiomaps_network/
OpenBirdMaps (2013) – Available at https://openbirdmaps.ro
OpenHerpMaps (2019) – Available at https://openherpmaps.ro
Pannon Bioregion Biotic Dataset (2012) – Available at https://dinpi.openbiomaps.org/projects/dinpi/, https://doi.org/10.18426/obm.23mj4em6bca0
Powers SM, Hampton SE (2019) Open science, reproducibility, and transparency in ecology. Ecol Appl. 29:e01822. https://doi.org/10.1002/eap.1822
Public Nestbox Breeding Data (2016) – Available at https://openbiomaps.org/projects/ public_nestbox_data/
Reichman O, Jones M, Schildhauer M (2011) Challenges and Opportunities of Open Data in Ecology. Science 331:703–705. https://doi.org/10.1126/science.1197962
Robertson T, Döring M, Guralnick R, Bloom D, Wieczorek J, Braak K, Otegui J, Russell L, Desmet P (2014) The GBIF integrated publishing toolkit: facilitating the efficient publishing of biodiversity data on the internet.PLoS One, 9(8), e102623
Sex Ratio in Vertebrates database (2017) – Available at https://openbiomaps.org/projects/sex_ratio_evolution/
Takács AA, Szilágyi G (2004) A Természetvédelmi információs rendszer kialakítása. Térinformatika (ISSN: 0864–8549) 16(4), 23–25
Tammaro AM, Matusiak KK, Sposito FA, Casarosa V (2019) Data Curator’s Roles and Responsibilities: An International Perspective. Libri 69:89–104. https://doi.org/10.1515/libri-2018-0090
Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M, Giovanni R, Robertson T, Vieglais D (2012) Darwin Core: an evolving community-developed biodiversity data standard.PloS one, 7(1), e29715
Wilkinson MD, Dumontier M, Aalbersberg IjJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone S-A, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B (2016) The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3:160018. https://doi.org/10.1038/sdata.2016.18
Acknowledgements
This research was supported by the European Union and the State of Hungary, co-financed by the European Social Fund within the framework of TÁMOP-4.2.4.A/ 2–11/1-2012-0001 ‘National Excellence Program’. This work benefited from services and resources provided by the EOSC-ACE project (receiving funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 101017567), with the dedicated support from the Advanced Computing and e-Science group at the Institute of Physics of Cantabria (IFCA-CSIC-UC). ZB was supported by the Thematic Excellence Programme (TKP2020-IKA-04) of the Ministry for Innovation and Technology in Hungary.
Funding
Open access funding provided by University of Debrecen.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare.
Additional information
Communicated by H. Babaie
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bán, M., Boné, G.M., Bérces, S. et al. OpenBioMaps – self-hosted data management platform and distributed service for biodiversity related data. Earth Sci Inform 15, 2007–2016 (2022). https://doi.org/10.1007/s12145-022-00818-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12145-022-00818-3