Abstract
Chrysophrys auratus (Australasian snapper) is one of the largest and most valuable finfish from capture fisheries in New Zealand, yet no cell lines from this species are reported in the scientific literature. Here, we describe a muscle-derived cell line initiated from the tail of a juvenile snapper which has been designated CAtmus1PFR (Chrysophrys auratus, tail muscle, Plant & Food Research). The cell line has been passaged over 100 times in 3 years and is considered immortal. Cells are reliant on serum supplementation for proliferation and exhibit a broad thermal profile comparable to the eurythermic nature of C. auratus in vivo. The impact of exogenous growth factors, including insulin-like growth factors I and II (IGF-I and IGF-II), basic fibroblast growth factor (bFGF), and transforming growth factor beta (TGFβ), on cell morphology and proliferation was investigated. Insulin-like growth factors acted as mitogens and had minimal effect on cell morphology. TGFβ exposure resulted in CAtmus1PFR exhibiting a myofibroblast morphology becoming enlarged with actin bundling. This differentiation was confirmed through the expression of smooth muscle actin (sma), an increase in type 1 collagen (col1a) expression, and a loss of motility. Expression of col1a and sma was decreased when cells were exposed to bFGF, and no actin bundling was observed. These data indicate that CAtmus1PFR may be myofibroblastic precursor cells descending from mesenchymal progenitor cells present in the tail muscle myosepta.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Australasian snapper (Chrysophrys auratus, formerly known as Pagrus auratus) is a marine teleost which is found in the coastal waters of Australia and New Zealand (Parsons et al. 2014). The species has economic significance in New Zealand, representing one of the largest capture fisheries in the country. In addition, snapper, or tāmure, have cultural significance to the indigenous people of New Zealand (Māori) as well as being a popular recreational fishery (Parsons et al. 2014). These factors have made snapper a prime candidate for aquaculture. A selective breeding programme to produce snapper strains that are adapted to domestication is ongoing in New Zealand (Ashton et al. 2019; Wellenreuther et al. 2019). Despite the significance of this species, there are no cell lines derived from C. auratus tissue, including muscle. As fish strains that exhibit more rapid muscle growth are desirable for commercial purposes (Olesen et al. 2003; Sandoval-Castillo et al. 2022), availability of muscle-derived cell lines could assist aquaculture of snapper through fundamental understanding of muscle growth, development, and repair in vitro, but would also provide a platform for controlled analysis of single or multiple parameters at a time, e.g. temperature, salinity, and nutrients. These types of analyses can be challenging when dealing with live organisms in complex aquatic environments. A well-characterised cell line from one species has the potential to act as a model system for other related species. There are also possible applications of cell lines beyond traditional aquaculture in the field of cellular agriculture (Rubio et al. 2019).
Fish skeletal muscle is composed mainly of muscle fibres which form through recruitment of new fibres (hyperplasia) or growth of existing fibres (hypertrophy) (Zimmerman and Lowery 1999; Ruparelia et al. 2020). The dimensions and distribution of fish muscle fibres, coupled with the extent of intramuscular fat, and the composition of intramuscular connective tissue are important factors which affect the final texture and quality of the fish flesh (Kiessling et al. 2006; Listrat et al. 2016). Despite this, description of specific cell types beyond fibre-forming stem/satellite cells isolated from fish embryos or skeletal muscle (Gignac et al. 2014; Peng et al. 2016; Sultan et al. 2021) is limited, particularly in comparison to mammalian systems where vasculature-associated stem cells including mesoangioblasts and pericytes have been described, in addition to interstitial cells such as fibro-adipogenic progenitor (FAP) cells and PW1-positive interstitial cells (PICs) (Cottle et al. 2017; Nassari et al. 2017; Rubenstein et al. 2020). The role of these and other progenitor cells of the myoseptum is essentially unexplored in fish muscle (Charvet et al. 2011; Bricard et al. 2014; Ruparelia et al. 2020).
The fish skeletal muscle invitrome (i.e. a grou** of cell lines around a theme or category (Bols et al. 2017)) is extremely limited. Currently, approximately 26 skeletal muscle cell lines from 20 fish species have been described in the scientific literature (Cellosaurus 2022). This number of species represents less than 0.04% of all known fish species (> 34,000) (www.fishbase.org). Of these cell lines, the majority are classified by cell morphology, mainly as fibroblastic or epithelial, with few progressing characterisation to describe a specific cell type (Fernandez et al. 1993; Zhao et al. 2003; Zhao and Lu 2006; Rougee et al. 2007; Lai et al. 2008). A key challenge in establishing well-characterised muscle (or any other tissue)-derived fish cell lines is the lack of resources such as validated, species-specific antibodies, detailed knowledge of cell lineages that exist in fish muscle, a lack of fundamental knowledge on the application of cell sorting technologies specifically for fish cells, and the huge breadth of species making transfer of technology and knowledge challenging (Rubio et al. 2019; Potter et al. 2020). In the absence of these resources, one way of determining a specific cell type is to investigate the genetic, physical, and biochemical responses of cells to various growth factors or cytokines, as their impacts on skeletal muscle development, homeostasis, and repair are reasonably well understood across species (Syverud et al. 2016, Vélez et al. 2017; Duran et al. 2022) . Fibroblast growth factor (FGF), transforming growth factor β (TGFβ), and insulin-like growth factors (IGFs) are some of the most influential cytokines involved in these processes (Dayton and Hathaway 1991; Syverud et al. 2016).
FGFs have been shown to promote proliferation of activated myogenic stem cells, suppressing their terminal differentiation (Dayton and Hathaway 1991; Fox and Swain 1993). In addition, FGFs have been shown to enhance the proliferation of fibroblasts and promote the formation of new blood vessels (Floss et al. 1997; Kastner et al. 2000). Myogenic stem cell differentiation is inhibited by TGFβ through the inhibition of myogenic regulators MyoD and myogenin (Liu et al. 2001). TGFβ regulates the phenotype and function of fibroblasts and plays a role in myofibroblast transdifferentiation (Krummel et al. 1988). This process promotes expression of extracellular matrix and profibrotic genes such as type 1 collagen (col1a) (Ignotz and Massagué 1986; Grande et al. 1997) and connective tissue growth factor (ctgf) (Leask et al. 2009; Tsai et al. 2018). In some situations, excessive TGFβ-induced deposition of extracellular matrix components can lead to fibrosis, a condition which severely impairs muscle function via reduced mobility and contractile function (Mann et al. 2011). Fibrosis in fish heart, liver, and muscle has been reported in vivo (Arana et al. 2014; Keen et al. 2015; Lin et al. 2022). IGFs stimulate nutrient uptake, enhance myoblast proliferation, and increase protein synthesis across a number of species (Negatu and Meier 1995; Rius-Francino et al. 2011; Castillo et al. 2004; Codina et al. 2008; Díaz et al. 2009; Gabillard et al. 2010; Montserrat et al. 2012). There is also evidence that exogenous addition of IGF-I to primary myocytes derived from gilthead seabream, a closely related species to the Australasian snapper, resulted in increased expression of myogenin, a transcriptional factor involved in myogenesis, indicating that IGF may also play a role in the differentiation process (Vélez et al. 2014).
Here we report on the first Chrysophrys auratus–derived cell line, among several others currently in development at Plant & Food Research (Nelson, New Zealand). This cell line will be identified as CAtmus1PFR (Chrysophrys auratus tail muscle 1 Plant & Food Research—species; tissue; institute affiliation). Our findings indicate that this cell line is a myofibroblastic progenitor cell derived from the non-myogenic line of mesodermal progenitor cells.
Materials and methods
Cell isolation and primary culture
A single healthy juvenile snapper (Chrysophrys auratus) bred at The New Zealand Institute for Plant and Food Research Limited was euthanised with an overdose of anaesthetic (60 ppm AQUI-S, Lower Hut, New Zealand). The fish weighed 1.45 g and was 4.5 cm in length from nose to peduncle (Fig. 1A). The specimen was kept on ice for 30 min before processing for cell culture. The fish was surface sterilised with 70% ethanol and approximately 1 cm3 of de-skinned muscle tissue was excised from the tail peduncle. The muscle segment was washed extensively in Hanks’ balanced salt solution (HBSS, Sigma Aldrich, St Louis, MO) supplemented with 100 units mL−1 penicillin and 100 µg mL−1 streptomycin (1 × P/S) (Sigma-Aldrich). Using sterile scalpel blades, the tissue was minced into small fragments which were further dissociated enzymatically by incubation with 1 mg mL−1 collagenase in HBSS at 18 °C for 4 h. The dissociated tissue fragment was centrifuged at 95 × g for 5 min at room temperature (Hettich 320R), after which the supernatant was removed and the remaining cell pellet resuspended in 2 mL Leibovitz’s L-15 (L-15, Gibco, Grand Island, NY) medium supplemented with 25% FBS (Gibco) and 1 × P/S. The resuspended cells were added to a 25-cm2 flask (Falcon, Corning, NY) which was incubated in the dark, in ambient air and temperature (approximately 18 °C). Twenty-four hours later, media containing unattached cells and debris was removed and fresh media added (L-15, 25% FBS, 1 × P/S). After about 6 weeks in culture, the original T25 flask reached confluency, and was passaged to a fresh flask as indicated below, and the serum concentration was reduced to 20%. Once the cells had been passaged four times, the serum concentration was reduced to 10%. Cells were monitored routinely using an inverted microscope (CKX53, Olympus, Tokyo, Japan).
Establishment of CAtmus1PFR muscle-derived cell line from Chrysophrys auratus. (A) Snapper from where CAtmus1PFR was derived. (B) Cells 9 d after isolation. (C) Early cell death. (D) Additional cell types (E–F) Cells with various morphologies after 1 month in culture. (G) Fibrous morphology of CAtmus1PFR when sub confluent. (H) Confluent morphology and (I) Confluent cells with sub-population of large rounded cells. * indicates same position on micrographs (B) and (C). All scale bars are 100 µm.
Cell line maintenance
Cells were routinely cultured in standard non-vented tissue culture flasks (Nunc EasYFlask, Thermo Fisher Scientific, Waltham, MA) at 18 °C, in the dark, in ambient air. L-15 medium supplemented with 10% FBS and 1 × P/S was used for routine culture. On reaching confluency, cells were detached from culture vessels by exposure to 1 × TrypLE Express (Gibco) for 5–6 min followed by centrifugation at 95 × g for 5 min. Spent medium was removed, and the cells were reseeded in fresh media at 0.133 mL media cm2- (i.e. 10 mL for a 75-cm2 flask). For cryopreservation, cells were pelleted (95 × g for 5 min) and resuspended in 0.5 mL L-15 media (10% FBS). An equal volume of freezing media (L-15 with 20% dimethyl sulfoxide (DMSO)) was added for a final cell concentration of 1 × 106 cells mL−1 and a final DMSO concentration of 10%. Cells were stored at − 80 °C for a minimum of 48 h before long-term storage in the vapour phase of liquid nitrogen.
Mycoplasma detection
Cells at passage 20, 28, and 48 h were cultured to confluency in the absence of antibiotics for 2 weeks. The presence of contaminating mycoplasma species was monitored using the Lookout® Mycoplasma PCR Detection Kit according to the manufacturer’s instructions (Sigma-Aldrich).
DNA barcoding
Cell culture provenance was verified by DNA barcoding using universal non-specific primers targeting conserved sequences of cytochrome c oxidase subunit I (COI) and 16 s rRNA, and comparison to amplicons of the same genes of gDNA extracted from whole fish. DNA was isolated from CAtmus1PFR using a Purelink™ Genomic DNA Extraction Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations. gDNA from a fin clip of C. auratus was extracted using the method described previously by Ashton et al. (2019). Briefly, a fresh fin clip was placed into chilled 96% ethanol followed by heating to 80 °C for 5 min. The sample was stored at − 20 °C until additional processing occurred. Approximately 20 mg of the fin clip was digested by incubation in extraction buffer (0.4 M NaCl, 10 mM Tris–HCl pH 8.0, 2 mM EDTA pH 8.0) with 2% SDS (final concentration) at 80 °C for 5 min followed by rapid cooling on ice. The sample was digested with proteinase K (400 µg mL−1 final concentration) for 1.5 h at 56 °C, after which the sample was centrifuged at 13,680 × g for 15 min. Following treatment with 5 M NaCl and centrifugation, RNAse A (100 µg mL−1) was added to remove contaminating RNA. DNA was precipitated with isopropanol overnight at − 20 °C. Subsequent ethanol washes followed by air-drying yielded purified gDNA.
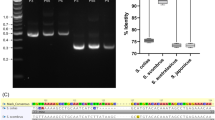
16 s rRNA and COI amplifications from gDNA extracted from the fin clip and cell line were performed using BIOTAQ™ DNA Polymerase (Meridian Bioscience, Cincinnati, OH) according to the manufacturer’s instructions. Forward and reverse primers (Table 1) were used to amplify approximately 750 bp of COI as follows: 94 °C for 4 min, followed by 30 cycles of 94 °C (50 s), 59 °C (50 s), and 72 °C (90 s), followed by 72 °C for 7 min. Approximately 1500 bp of the 16 s rRNA gene (~ 1500 bp) was amplified in a similar fashion but with a 54 °C annealing temperature. PCR products run on a 1% agarose gel were extracted using a Nucleospin Gel and PCR Clean-up kit (Machery Nagel, Duren, Germany) (Fig. S2). Sanger sequencing of gel-extracted amplicons was carried out by Genetic Analysis Services (University of Otago, New Zealand). Sequence reads from amplified COI and 16 s rRNA genes from CAtmus1PFR gDNA and gDNA extracted from the whole fish were aligned using BioEdit. Where errors became frequent, sequences were truncated. Chromatograms were scanned for miscalls and corrected where deemed appropriate. Using the forward and reverse amplification primers as previously, a consensus sequence of 622 bp resulted for COI. For 16 s rRNA, a sequencing primer coupled with the previously used forward primer was used to generate a 694 bp consensus sequence.
Effects of FBS, basal media, and temperature on CAtmus1PFR cell growth
Cells (1 × 104) were cultured in 24-well plates containing 1 mL of standard culture media (L-15, 1 × P/S, 10% FBS) and incubated at 18 °C for 24 h to allow cell attachment to occur. Following this period, cells were challenged with varying FBS concentrations (0%, 5%, 10%, 15%, 20%, or 25%) or different basal media formulations (L-15, CO2 Independent Medium (CIM, Gibco), Medium 199, Hanks’ Balanced Salts (M199-HBSS, Gibco), MEM, Hanks’ Balanced Salts (MEM-HBSS, Gibco) supplemented with 10% FBS and 1 × P/S. At 2, 7, 10, and 14 days post inoculation, cells were detached in 1 × TrypLE Express and counted using a haemocytometer. The impact of temperature on CAtmus1PFR cells was examined by seeding each well of 24-well plates with 1 × 104 cells and incubating immediately at various temperatures (4 °C, 12 °C, 18 °C, 24 °C, 30 °C, 32 °C). Cell number was determined as above. Data shown in Fig. 2 represent data from three biological replicates carried out on different days. Each biological replicate had three technical replicates.
Impact of growth factors on cell growth
Pilot experiments confirmed the correlation between cell number and measured cell metabolic activity through use of an MTT assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide, Invitrogen; data not shown), and so, this method was employed to monitor the impact of basic fibroblast growth factor (bFGF, human recombinant, Catalogue No. PHG0266, Thermo Fisher), transforming growth factor beta 1 (TGFβ1, human recombinant, Catalogue No. PHG9204, Thermo Fisher), insulin-like growth factor-1 (IGF-I, human recombinant, Catalogue No. PHG0071, Thermo Fisher), and insulin-like growth factor-2 (IGF-II, human recombinant, Catalogue No. PHG0084, Thermo Fisher) on cell proliferation.
Twenty-four-well plates were seeded at 1 × 104 cells mL−1 in standard culture media (L-15, 10% FBS, 1 × P/S) supplemented with 1 or 100 ng mL−1 of a single growth factor. All plates were incubated in the dark at 18 °C in ambient air. At 1, 4, 7, and 10 days, an MTT assay was carried out as described previously. Briefly, wells were washed twice with 1 mL phosphate-buffered saline (PBS, Thermo Fisher), followed by addition of 0.4 mL serum-free media supplemented with 0.1 mL MTT (5 mg mL−1). Cells were incubated for 70 min at room temperature, after which 0.5 mL DMSO was added to each well. Cells were incubated with agitation for 10 min followed by gentle trituration. A 0.9 mL sample from each well was transferred to a cuvette where the absorbance was measured at 600 nm (Evolution 220 UV–Visible Spectrophotometer, Thermo Fisher). The spectrophotometer was blanked using serum-free L-15, MTT, and DMSO.
RNA extraction and qPCR
Cells were seeded at 1 × 104 cells mL−1 in media (L-15, 10% FBS, 1 × P/S) supplemented with 1 or 100 ng mL−1 growth factors (bFGF, IGF-I, IGF-II, or TGFβ1). After 4 days in culture, cells were washed twice in 1 mL dPBS and RNA was extracted and combined from four wells for each condition using a GENEzol TriRNA Pure Kit (Geneaid, New Taipei City, Taiwan) as per the manufacturer’s recommendations. An on-column DNase treatment was carried out to remove genomic DNA. Complementary DNA (cDNA) was synthesised from 500 ng of RNA using Superscript VILO (Invitrogen) in a 20 µL reaction volume. qPCR primers were designed based on the gilthead seabream (Sparus aurata) which is closely related to C. auratus. Database accession numbers of each target and reference genes are identified in Table 2. All primers were designed to span an exon-exon junction and should, therefore, not detect genomic DNA, sequences of which are shown in Table 2. Primer efficiencies were calculated to be between 91 and 98% (Table 2). Real-time PCR was performed using a Quant Studio 6 Flex instrument (Thermo Fisher). Reactions consisted of 0.5 µL forward and reverse primers (10 µM), 8 µL TB Green® (Takara Bio, Shiga, Japan), 6 µL UP H2O, and 1 µL cDNA for a total reaction volume of 16 µL.
Elongation factor 1a (elfa) was used as reference gene and the threshold cycle (CT) values were between 13 and 14. CT values for col1A, dcn, sma, and vim were ~ 16, ~ 23, ~ 16, and ~ 23 in the control conditions respectively. The relative fold change in gene expression was calculated using the delta delta CT method (2−ΔΔCT) as described by Livak and Schmittgen (2001).
F-actin labelling
Cells were seeded at a density of 1 × 104 cells/well in a 24-well plate in the presence or absence of 100 ng mL−1 bFGF or TGFβ. After 4 days in culture under standard conditions, wells were washed in dPBS and cells fixed in 4% paraformaldehyde for 30 min at room temperature. Cells were permeabilised in 0.4% Triton-X-100 in dPBS for 20 min followed by treatment with Rhodamine phalloidin (Invitrogen) in 0.1% BSA in dPBS for 1 h. Cells were washed and imaged using fluorescence microscopy (CKX53, Olympus ).
Migration assay
Cells (1.2 × 103 mL−1) suspended in L-15 growth medium were seeded in 0.07 mL volume in each side of a two-well migration chamber (IBIDI, Bavaria, Germany). Growth factors (bFGF or TGFβ) were added to a final concentration of 1 or 100 ng mL−1. Chambers were incubated in ambient air and temperature, and the media refreshed on day 4. On day 7, the migration chamber was removed, leaving a 0.5-cm cell-free gap between cell populations. Debris was removed by washing in 1 mL dPBS. Fresh growth factor containing growth media was added, and the plates were imaged immediately using an inverted microscope. Subsequent images of the cell-free gap were recorded at 6 and 24 h post removal from the migration chamber. The gap in a 1500 × 450 pixel (equivalent to ~ 3.5 mm × ~ 1 mm) section from each condition was measured using ImageJ software. Each migration assay, of which there were two biological replicates, had triplicate technical replicates.
Animal handling
All research carried out in this study was reviewed and approved by the Animal Ethics Committee of Nelson Marlborough Institute of Technology in New Zealand (Application number AEC2019-PFR-01).
Results
CAtmus1PFR is a continuous cell line derived from C. auratus tail skeletal muscle
Collagenase treatment of a muscle segment derived from a juvenile C. auratus (Fig. 1A) resulted in a preparation with a significant amount of debris. This was not consistent with microbial contamination, but rather appeared to be fragments of degraded muscle fibres (Fig. 1B, C). By day 9, small colonies of replicating cells with fibroblastic or spindle morphologies were observed (Fig. 1B). Many of these early cell types died off and were replaced with other proliferative cells with varying morphologies (Fig. 1C, D). After 1 month in culture, as the primary culture neared confluency, a heterogenous population of cells was evident with a number of different cell morphologies (Fig. 1E, F). Following continuous passage, a single cell type emerged which had long spindle-like processes when culturing (Fig. 1G). However, on reaching confluency, a subpopulation of cells with a larger, more rounded morphology developed. Cells were successfully cryopreserved and resuscitated in L-15 with 10% FBS and yielded approximately 80–90% cell viability. No morphological changes were observed following resuscitation. At the time of publication, the cell line has been passaged 100 times over a 3-yr period. The line is assumed continuous through spontaneous immortalisation, as is common for finfish cell lines, and has been designated CAtmus1PFR (Chrysophrys auratus, tail muscle, Plant and Food Research). The absence of Mycoplasma species was confirmed through PCR (Fig. S1). The species origin was verified as Chrysophrys auratus by 100% homology between 16s rRNA and COI gene sequence from CAtmus1PFR genomic DNA to gDNA extracted from a C. auratus fin clip.
Temperature, serum concentration, and basal media formulation impact proliferation of CAtmus1PFR
The thermal tolerance of CAtmus1PFR to temperatures ranging from 4 to 32 °C was investigated (Fig. 2A). At hypothermic conditions (4 °C), the cells completely detached from the culture vessel after 2 d in culture. At 12 °C, the cells proliferated slowly, and, by day 14, the final numbers were approximately half that of normothermic conditions (18 °C). At 32 °C (hyperthermic), the cells were unable to replicate, but remained attached to the tissue culture vessel. Remaining cells were enlarged in size compared with cells cultured at normothermic conditions (18 °C). Temperatures ranging from 18 to 30 °C provided conditions for comparable growth of CAtmus1PFR cells with no variation in cell morphology (Fig. 2A).
In the absence of serum, the number of viable cells decreased over time; by day 14, a small number of cells remained with an elongated morphology (Fig. 2B and S3). Decreasing FBS concentration from the standard 10 to 5% resulted in a lower proliferative rate. Conversely, increasing the serum concentration resulted in a dose-dependent increase in cell number over a 14-d period (Fig. 2B). In media supplemented with 25% FBS, the cell number after 2 wk in culture was more than double that observed with 10% FBS (Fig. 2B). There was no impact on cell morphology with FBS concentrations between 5 and 25%.
CAtmus1PFR is routinely cultured in L-15 media which allows the cells to be cultured without the need for additional CO2. Alternative buffered media were investigated for their impact on proliferation of CAtmus1PFR cells (Fig. 2C). After 7 days in culture, proliferation of CAtmus1PFR in M199 was significantly greater than that observed in three other basal media preparations (L-15, CO2-independent medium [CIM], and MEM HBSS). By day 10, the maximum cell number had been reached in M199 (~ 1.1 × 105), and this was matched by CIM. Interestingly, cells culturing in L-15 and MEM reached maximal number at 7 d (~ 6 × 104), and did not show further growth even with fresh media replacements. Final cell numbers in MEM and L-15 were significantly lower than in CIM and M199.
Growth factors affect proliferation and morphology of CAtmus1PFR
At 1 ng mL−1, insulin-like growth factors I and II (IGF-I, IGF-II) had no impact on cell number as measured by MTT assay; however, at 100 ng mL−1, both IGF-I and IGF-II resulted in increased cell numbers (Fig. 3B, D).
Impact of growth factors on morphology and replication of CAtmus1PFR. (A) Micrographs showing pleiomorphic phenotype in control condition (L-15 media supplemented with 10% FBS and 1 × penicillin streptomycin) when confluent. Impact of IGF-I (B, C), IGF-II (D, E), TGFβ1 (E, F), and bFGF (G, H) on CAtmus1PFR proliferation and morphology at 1 and 100 ng mL−1 (n = 3). Error bars = means ± standard deviation. All scale bars are 100 µm.
TGFβ1 had no impact on cell numbers, but had a significant impact on the cell morphology (Fig. 3F, G). The cell size increased and appeared to be similar to the subpopulation of cells observed under standard culture conditions (Fig. 3A). bFGF had minimal impact on proliferation of CAtmus1PFR but maintained all the cells in a single morphology (Fig. 3H, I) similar to those observed in the control condition. The large rounded cells, which formed in the control condition (Fig. 3A), did not form in the presence of bFGF (Fig. 3I).
CAtmus1PFR treated with TGFβ have myofibroblast properties
The change in morphology of CAtmus1PFR following treatment with TGFβ1 and bFGF suggested differentiation of the cell. Previous data have demonstrated that TGFβ1 can result in the differentiation of precursor cells to myofibroblasts. These differentiated cells exhibit changes in cell shape, modified cytoskeletal features including the formation of actin bundles, and increased deposition of extracellular matrix components such as collagen. bFGF has been shown to have an opposing effect, causing downregulation of myofibroblastic features and encouraging a more fibroblastic phenotype. We therefore sought to determine if CAtmus1PFR was a myofibroblast progenitor cell, and gave rise to myofibroblasts following TGFβ treatment, and whether bFGF had an opposing effect on this process.
Myofibroblasts are associated in vivo with enhanced deposition of collagen. In TGFβ1-treated cells, a 2.5- to threefold increase in expression of col1a mRNA at 1 and 100 ng mL−1, respectively, was observed (Fig. 4A). Conversely, exposing cells to 1 or 100 ng mL−1 bFGF resulted in a significant decrease in expression. The expression of decorin mRNA, from a gene encoding a proteoglycan extracellular matrix (ECM) component, was also increased in the presence of TGFβ1 and decreased where the cells were cultured with bFGF (Fig. 4B).
Impact of growth factors on expression of genes associated with extracellular matrix components. Change in expression of (A) type 1 collagen 1 (col1A) and (B) decorin (dcn) following 4 d of treatment with IGF-I, IGF-II, TGFβ1, or bFGF at 1 and 100 ng mL−1 relative to control condition (L-15, 10% FBS, 1 × P/S) (n = 3). Error bars = means ± standard deviation. *P < 0.05 by one-way ANOVA followed by Tukey’s least Significant Difference (α = 0.05).
Myofibroblasts have been shown to exhibit actin bundling, which can be visualised with the fluorescent probe phalloidin, which binds to filamentous actin (F-actin). Under standard culture conditions (no growth factors), large cells with actin bundling and smaller cells without bundling were evident after 7 d in culture (Fig. 5B). In the presence of bFGF, no actin bundling was observed in any of the cells and the size and morphology remained unchanged (Fig. 5C, D). In the presence of TGFβ, cells exhibited significant actin bundling and large cell size by 4 d in culture (Fig. 5E). By day 7, the cells formed a mesh of cells with significant actin bundling (Fig. 5F). An additional cytoskeletal marker, the intermediate filament protein, vimentin, which is known to be expressed by myofibroblasts, was also upregulated following TGFβ1 treatment of CAtmus1PFR cells and downregulated in the presence of bFGF, although the changes were not statistically significant (Fig. 5G).
Effects of growth factors on the cytoskeletal structure of CAtmus1PFR cells. Cells 4 and 7 d in culture under standard conditions (A, B), or in the presence of 100 ng mL−1 bFGF (C, D) or TGFβ1 (E, F). (G) Expression of vimentin after 4 d exposure to TGFβ1 and bFGF at 1 and 100 ng mL−1 in comparison to control unexposed cells. Error bars = means ± standard deviation. All scale bars are 100 µm.
Expression of smooth muscle actin (SMA) is generally indicative of myofibroblast differentiation, and, when exposed to TGFβ1, expression of this gene increases in CAtmus1PFR (Fig. 6A). A large variation in this increased expression was observed across biological replicates (changes between 2- and eightfold). It is plausible that the status of the seeding culture plays an important role in the expression profile of sma, as cultures which have a proportion of cells already differentiated to myofibroblasts may result in an increased expression of sma in the cell population. Conversely, bFGF-treated cells decrease expression of sma mRNA (Fig. 6A). SMA protein has previously been shown to be involved with reducing the migratory capabilities of cells. To further investigate the migratory capabilities of growth factor–treated cells, a wound-migration assay was carried out. Mobility of cells treated with TGFβ1 was almost completely abolished at 100 ng mL−1, and was somewhat retarded at 1 ng mL−1 (Fig. 6C, D). Untreated and cells treated with bFGF migrated into and closed the gap within 24–36 h (Fig. 6C, D).
Effects of growth factors on expression of smooth muscle actin (sma) and on cell migration. (A) Expression of sma after 4 d exposure to TGFβ and bFGF at 1 and 100 ng mL−1 in comparison to control unexposed cells. Symbols represent fold change from 4 independent biological replicates for each condition. Bars represent the mean value for each condition. (B) Measurement of gap size at initial ‘wounding’ and 24 h later for cells cultured in standard culture media (L-15, 10% FBS, 1 × P/S), or media supplemented with bFGF or TGFβ at 1 and 100 ng mL−1. Values represent means and standard deviations of triplicate biological replicates, each with two or three technical replicates. (D) Micrographs demonstrating migration of CAtmus1PFR cultured in control and growth factor-supplemented media over 24 h. Scale bars are 100 µm.
Discussion
Here we describe the growth characteristics and differentiation capacity of a muscle-derived myofibroblastic precursor cell line from the tail peduncle muscle of Australasian snapper. This is the first reported cell line for C. auratus, and it joins 26 other cell lines derived from fish skeletal muscle reported in the scientific literature. Of the previously reported lines, the majority are described as being fibroblastic according to their morphology. In many cases, these have been developed as in vitro systems for monitoring and control of infectious disease in aquaculture (Middlebrooks et al. 1981; Zhao et al. 2003; Zhao and Lu 2006; Lai et al. 2008; Wang et al. 2020). Despite the emphasis on fibroblasts, skeletal muscle is a complex environment with many cell types, each playing an important role in the development, repair, and homeostasis of muscle tissue (Joe et al. 2010; Uezumi et al. 2011; Heredia et al. 2013; Giordani et al. 2019). Additionally, the ECM supporting these cells provides both the physical and biochemical environments in which these cells reside (Csapo et al. 2020). Satellite cells, long considered muscle progenitor cells, are abundant in fish (Johnston 2006; Ruparelia et al. 2020), and have been shown to undergo dichotomic differentiation into myoblastic and non-myoblastic lineages (Shefer et al. 2004). The myoblastic lineage leads to myogenic development, whereas the alternative leads to a mesenchymal pathway giving rise to cells which reside in the connective tissue of muscle. Additionally, mesenchymal stem cells distinct from satellite cells have also been described to occur in skeletal muscle tissues. Work is ongoing to determine the specific stem cell origins that has given rise to CAtmus1PFR.
Fish cell lines have been described as having a proliferation zone with an optimum temperature where cells replicate more rapidly (Bols et al. 1992). In the case of CAtmus1PFR, there was a broad optimal temperature where the cells had a similar proliferation capacity (between 18 and 30 °C). This is in comparison to a number of other cell lines where a single temperature is identified as being optimal. A number of rainbow trout cell lines (RTE, RTE-2, RTG-2, RTH-149, and RTT) share an optimal temperature of 20 °C, with decreased proliferation at 5 °C above or below this temperature (Bols et al. 1992). The broad temperature tolerance of CAtmus1PFR may be due to the eurythermal nature of snapper (Parsons et al. 2014), suggesting cell lines from this species represent the in vivo nature of the fish. It would be of interest to determine if the effect of temperature on the gene expression profile of CAtmus1PFR mirrors that observed in snapper in vivo, where pathways associated with metabolism, cell regulation, and cell signalling were implicated (Wellenreuther et al. 2019).
The specific nutritional requirements of skeletal muscle-derived fish cell lines are undefined. Basal media formulations developed for mammalian cells supplemented with mammalian serum as a source of growth factors, lipids, and additional nutrients are routinely used (Bols and Lee 1994). Proliferation of many muscle-derived fish cell lines is positively affected by increasing serum concentrations (Zhao et al. 2003; Zhao and Lu 2006; Rougee et al. 2007; Wang et al. 2020), and this is similar to that observed with CAtmus1PFR. Interestingly, different fish species have different tolerances to basal media formulations (Fernandez et al. 1993; Wang et al. 2003; Zhao and Lu 2006). Here we have demonstrated CAtmus1PFR is capable of enhanced replication in M199 and CIM media formulations; however, as the impact(s) these media had on the biochemical and physical responses of cells was not determined, we continued to culture the cell line in the medium in which it was established (L-15). It is unclear at this stage whether media preferences are determined by the donor fish species, the donor tissue, or a specific cell type. A comparison of requirements between these variables may provide additional information when it comes to understanding the development and growth of fish muscle for applications in both traditional and cellular aquaculture (Rubio et al. 2019).
In fish skeletal muscle, mesenchymal progenitor cells are believed to give rise to myogenic and non-myogenic lineages. The myogenic lineage leads to the formation of skeletal muscle per se, while the non-myogenic lineage leads to myofibroblast generation and is also proposed to form the resident fibroblasts, fibro/adipogenic progenitor cells, pericytes, and stem cells (Paylor et al. 2011). The impacts of various growth factors on fish myogenic cells have been reasonably understood, but their effects on the non-myogenic lineage less so. Here we provide evidence that CAtmus1PFR is a non-myogenic mesenchymal progenitor cell that is capable of differentiation to a myofibroblast.
Under normal physiological conditions, many components of the ECM, including type 1 collagen, are produced by muscle-resident fibroblasts. In situations when muscle tissue becomes damaged, myofibroblasts are recruited. These specialised cells increase deposition of collagen, which aids in the repair of muscle tissue, but where the action of these cells is not resolved, a thickening or scarring of the tissue can occur (fibrosis). Decorin, another component of the ECM, prevents the formation of fibrosis by binding to TGFβ, yet there is evidence that myofibroblasts express full-length biologically active decorin, which essentially contradicts the role myofibroblasts play in the formation of fibrosis in vivo (Honda and Munakata 2004). The exact reason for this is not clear but it has been proposed that decorin may play different roles as fibrosis progresses (Klingberg et al. 2013). It would be interesting to monitor decorin expression over time in CAtmus1PFR and determine if expression of decorin changes.
TGFβ is a known stimulant of myofibroblast differentiation and fibrosis (reviewed in Frangogiannis 2020). The differentiation process increases expression of type 1 collagen (col1a), and induces the expression of alpha-smooth muscle actin (sma) expression in myofibroblasts (Desmoulière et al. 1993), which may participate in the formation of stress fibres, cell contractility, and cell spreading characterising the myofibroblasts phenotype. TGFβ1 also contributes to the upregulation of vimentin expression while blocking myogenesis (Wu et al. 2007) and reducing migration. CAtmus1PFR responded similarly to TGFβ1, and these cells are thus prime for studying in vitro fibrosis events.
In vivo, increased deposition of collagen by cardiac myofibroblasts has been shown to play a role in the remodelling of the ECM of the heart in rainbow trout in response to cold acclimation (Johnston and Gillis 2018). This reactive fibrosis leads to a stiffening of the heart, which provides support for the increasing muscle mass of the fish. This has implications in aquaculture, where fish strains are specifically bred for their ability to increase muscle mass rapidly (Olesen et al. 2003). Indeed, perivertebral fibrosis, presumably due to the action of myofibroblasts, has been reported in farmed Chinook salmon which has led to spinal deformities, thus reducing the value of this commercially important species. The exact cause of these deformities is as yet unknown, but a number of factors have been proposed, including temperature, toxicities, trauma, and vitamin deficiencies (Munday et al. 2016). Indeed, the mycotoxin, aflatoxin B1, has been shown to cause liver fibrosis in trout (Arana et al. 2014), so it would be of interest to determine if this, and other toxins, in addition to compounding effects such as temperature stress, affected fibrosis in an in vitro model. Having a model of fibrosis is a useful tool, not only for understanding possible causes of fibrosis, but also for the development of prevention and rescue strategies, assuming the process is dynamic and, therefore, reversible. While myofibroblast precursor cells have previously been isolated and cultured from heart tissue of rainbow trout (Johnston and Gillis 2018), this is the first time a precursor cell capable of in vitro differentiation has been isolated from fish muscle. Beyond applications in traditional aquaculture, availability of this non-myogenic lineage from snapper tail muscle could also be relevant to the blossoming cellular aquaculture industry where the main focus has been in culturing myogenic cells, but as fish meat texture and structure are heavily influenced by the myoseptum and ECM, myofibroblasts and their precursors may also prove essential for this technology.
References
Arana S, Alves VAF, Sabino M, Tabata YA, Nonogaki S, Zaidan-Dagli ML, Hernandez-Blazquez FJ (2014) Immunohistochemical evidence for myofibroblast-like cells associated with liver injury induced by Aflatoxin B1 in rainbow trout (Oncorhynchus mykiss). J Comp Pathol 150:258–265. https://doi.org/10.1016/j.jcpa.2013.07.003
Ashton DT, Hilario E, Jaksons P, Ritchie PA, Wellenreuther M (2019) Genetic diversity and heritability of economically important traits in captive Australasian snapper (Chrysophrys auratus). Aquaculture 505:190–198. https://doi.org/10.1016/j.aquaculture.2019.02.034
Bols NC, Lee LEJ (1994) Cell lines: availability, propagation and isolation. In: Hochachka PW, Mommsen TP (eds) Biochemistry and molecular biology of fishes. Elsevier Science Publishers, pp 145–159. https://doi.org/10.1016/B978-0-444-82033-4.50019-2
Bols NC, Mosser DD, Steels GB (1992) Temperature studies and recent advances with fish cells in vitro. Comp Biochem Phys A 103:1–14. https://doi.org/10.1016/0300-9629(92)90235-I
Bols NC, Pham PH, Dayeh VR, Lee LEJ (2017) Invitromatics, invitrome, and invitroomics: introduction of three new terms for in vitro biology and illustration of their use with the cell lines from rainbow trout. In Vitro Cell Dev Biol Anim 53:383–405. https://doi.org/10.1007/s11626-017-0142-5
Bricard Y, Ralliere C, Lebret V, Lefevre F, Rescan PY (2014) Early fish myoseptal cells: insights from the trout and relationships with amniote axial tenocytes. Plos One 9(3):e91876
Castillo J, Codina M, Martinez ML, Navarro I, Gutierrez J (2004) Metabolic and mitogenic effects of IGF-I and insulin on muscle cells of rainbow trout. Am J Physiol Regul Integr Comp Physiol 286:R935–941. https://doi.org/10.1152/ajpregu.00459.2003
Cellosaurus (2022) A knowledge resource on cell lines. Available via https://web.expasy.org/cellosaurus/. Accessed 01 Sept 2022
Charvet B, Malbouyres M, Pagnon-Minot A, Ruggiero F, Le Guellec D (2011) Development of the zebrafish myoseptum with emphasis on the myotendinous junction. Cell Tissue Res 346:439–449. https://doi.org/10.1007/s00441-011-1266-7
Codina M, Garcia de la serrana D, Sanchez-Gurmaches J, Montserrat N, Chistyakova O, Navarro I, Gutierrez J (2008) Metabolic and mitogenic effects of IGF-II in rainbow trout (Oncorhynchus mykiss) myocytes in culture and the role of IGF-II in the PI3K/Akt and MAPK signalling pathways. Gen Comp Endocrinol 157:116–124. https://doi.org/10.1016/j.ygcen.2008.04.009
Cottle BJ, Lewis FC, Shone V, Ellison-Hughes GM (2017) Skeletal muscle-derived interstitial progenitor cells (PICs) display stem cell properties, being clonogenic, self-renewing, and multi-potent in vitro and in vivo. Stem Cell Res Ther 8(158). https://doi.org/10.1186/s13287-017-0612-4
Csapo R, Gumpenberger M, Wessner B (2020) Skeletal muscle extracellular matrix - what do we know about its composition, regulation, and physiological roles? A Narrative Review Front Physiol 11:253. https://doi.org/10.3389/fphys.2020.00253
Dayton WR, Hathaway MR (1991) Myogenic cell-proliferation and differentiation. Poult Sci 70:1815–1822. https://doi.org/10.3382/ps.0701815
Desmoulière A, Geinoz A, Gabbiani F, Gabbiani G (1993) Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol 122:103–111. https://doi.org/10.1083/jcb.122.1.103
Díaz M, Vraskou Y, Gutiérrez J, Planas JV (2009) Expression of rainbow trout glucose transporters GLUT1 and GLUT4 during in vitro muscle cell differentiation and regulation by insulin and IGF-I. Am J Physiol Regul Integr Comp Physiol 296:R794–R800. https://doi.org/10.1152/ajpregu.90673.2008
Duran BOS, Zanella BTT, Perez ES, Mareco EA, Blasco J, Dal-Pai-Silva M, de la Serrana DG (2022) Amino acids and IGF1 regulation of fish muscle growth revealed by transcriptome and micrornaome integrative analyses of Pacu (Piaractus mesopotamicus) Myotubes. Int J Mol Sci 23(3):1180. https://doi.org/10.3390/ijms23031180
Fernandez RD, Yoshimizu M, Ezura Y, Kimura T, Fernandez RD, Mamoru Y, Yoshio E, Takahisa K, Fenandez RD (1993) Comparative growth response of fish cell lines in different media, temperatures, and sodium chloride concentrations. Gyobyo Kenkyu 28:27–34. https://doi.org/10.3147/jsfp.28.27
FishBase (2022) http://www.fishbase.org. Cited 01 September 2022
Floss T, Arnold HH, Braun T (1997) A role for FGF-6 in skeletal muscle regeneration. Genes Dev 11:2040–2051. https://doi.org/10.1101/gad.11.16.2040
Fox JC, Swain JL (1993) Auto and transactivation of Fgf expression - potential mechanism for regulation of myogenic differentiation. In Vitro Cell Dev-An 29a:228–230. https://doi.org/10.1007/BF02634188
Frangogiannis N (2020) Transforming growth factor-β in tissue fibrosis. J Exp Med 217:e20190103. https://doi.org/10.1084/jem.20190103
Gabillard JC, Sabin N, Paboeuf G (2010) In vitro characterization of proliferation and differentiation of trout satellite cells. Cell Tissue Res 342:471–477. https://doi.org/10.1007/s00441-010-1071-8
Gignac SJ, Vo NTK, Mikhaeil MS, Alexander JAN, MacLatchy DL, Schulte PM, Lee LEJ (2014) Derivation of a continuous myogenic cell culture from an embryo of common killifish, Fundulus heteroclitus. Comp Biochem Physiol A Mol Integr Physiol 175:15–27. https://doi.org/10.1016/j.cbpa.2014.05.002
Giordani L, He GJ, Negroni E, Sakai H, Law JYC, Siu MM, Wan R, Corneau A, Tajbakhsh S, Cheung TH, Le Grand F (2019) High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol Cell 74:609-621.e606. https://doi.org/10.1016/j.molcel.2019.02.026
Grande JP, Melder DC, Zinsmeister AR (1997) Modulation of collagen gene expression by cytokines: stimulatory effect of transforming growth factor-beta1, with divergent effects of epidermal growth factor and tumor necrosis factor-alpha on collagen type I and collagen type IV. J Lab Clin Med 130:476–486. https://doi.org/10.1016/s0022-2143(97)90124-4
Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, Rando TA, Chawla A (2013) Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell 153:376–388. https://doi.org/10.1016/j.cell.2013.02.053
Honda E, Munakata H (2004) Purification and characterization of decorin from the culture media of MRC-5 cells. Int J Biochem Cell Biol 36:1635–1644. https://doi.org/10.1016/j.biocel.2004.01.023
Ignotz RA, Massagué J (1986) Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem 261:4337–4345. https://doi.org/10.1016/S0021-9258(17)35666-1
Joe AWB, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FMV (2010) Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol 12:153–163. https://doi.org/10.1038/ncb2015
Johnston EF, Gillis TE (2018) Transforming growth factor-beta1 induces differentiation of rainbow trout (Oncorhynchus mykiss) cardiac fibroblasts into myofibroblasts. J Exp Biol 221(24):jeb189167. https://doi.org/10.1242/jeb.189167
Johnston IA (2006) Environment and plasticity of myogenesis in teleost fish. J Exp Biol 209:2249–2264. https://doi.org/10.1242/jeb.02153
Kastner S, Elias MC, Rivera AJ, Yablonka-Reuveni Z (2000) Gene expression patterns of the fibroblast growth factors and their receptors during myogenesis of rat satellite cells. J Histochem Cytochem 48:1079–1096. https://doi.org/10.1177/002215540004800805
Keen AN, Fenna AJ, McConnell JC, Sherratt MJ, Gardner P, Shiels HA (2015) The dynamic nature of hypertrophic and fibrotic remodeling of the fish ventricle. Front Physiol 6:427. https://doi.org/10.3389/fphys.2015.00427
Kiessling A, Ruohonen K, Bjørnevik M (2006) Muscle fibre growth and quality in fish. Archives of Animal Breeding 49:137–146
Klingberg F, Hinz B, White ES (2013) The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol 229:298–309. https://doi.org/10.1002/path.4104
Kochzius M, Nolte M, Weber H, Silkenbeumer N, Hjorleifsdottir S, Hreggvidsson GO, Marteinsson V, Kappel K, Planes S, Tinti F, Magoulas A, Vazquez EG, Turan C, Hervet C, Falgueras DC, Antoniou A, Landi M, Blohm D (2008) DNA microarrays for identifying fishes. Mar Biotechnol 10:207–217. https://doi.org/10.1007/s10126-007-9068-3
Kochzius M, Seidel C, Antoniou A, Botla SK, Campo D, Cariani A, Vazquez EG, Hauschild J, Hervet C, Hjorleifsdottir S, Hreggvidsson G, Kappel K, Landi M, Magoulas A, Marteinsson V, Nolte M, Planes S, Tinti F, Turan C, Venugopal MN, Weber H, Blohm D (2010) Identifying fishes through DNA barcodes and microarrays. Plos One 5(9):e12620. https://doi.org/10.1371/journal.pone.0012620
Krummel TM, Michna BA, Thomas BL, Sporn MB, Nelson JM, Salzberg AM, Cohen IK, Diegelmann RF (1988) Transforming growth factor-beta (Tgf-Beta) induces fibrosis in a fetal wound model. J Pediatr Surg 23:647–652. https://doi.org/10.1016/S0022-3468(88)80638-9
Lai YS, Chiou PP, Chen WJ, Chen YC, Chen CW, Chiu IS, Chen SD, Cheng YH, Chang CY (2008) Characterization of apoptosis induced by grouper iridovirus in two newly established cell lines from barramundi, Lates calcarifer (Bloch). J Fish Dis 31:825–834. https://doi.org/10.1111/j.1365-2761.2008.00957.x
Leask A, Parapuram SK, Shi-Wen X, Abraham DJ (2009) Connective tissue growth factor (CTGF, CCN2) gene regulation: a potent clinical bio-marker of fibroproliferative disease? J Cell Commun Signal 3:89–94. https://doi.org/10.1007/s12079-009-0037-7
Lin F, Lin JL, Liu X, Yuan YY, Liu GQ, Ye XK (2022) Effects of temperature on muscle growth and collagen deposition in zebrafish (Danio rerio). Aquacult Rep 22:100952 https://doi.org/10.1016/j.aqrep.2021.100952
Listrat A, Lebret B, Louveau I, Astruc T, Bonnet M, Lefaucheur L, Picard B, Bugeon J (2016) How muscle structure and composition influence meat and flesh quality. Sci World J 2016:ID3182746. https://doi.org/10.1155/2016/3182746
Liu D, Black BL, Derynck R (2001) TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev 15:2950–2966. https://doi.org/10.1101/gad.925901
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mann CJ, Perdiguero E, Kharraz Y, Aguilar S, Pessina P, Serrano AL, Muñoz-Cánoves P (2011) Aberrant repair and fibrosis development in skeletal muscle. Skeletal Muscle 1:21. https://doi.org/10.1186/2044-5040-1-21
Middlebrooks BL, Stout DL, Ellender RD, Safford S (1981) Fish cell lines: two new cell lines derived from explants of trunk musculature of Cynoscion arenarius. In Vitro 17:427–430. https://doi.org/10.1007/BF02626743
Montserrat N, Capilla E, Navarro I, Gutiérrez J (2012) Metabolic effects of insulin and IGFs on gilthead sea bream (Sparus aurata) muscle cells. Front Endocrinol 3:55. https://doi.org/10.3389/fendo.2012.00055
Munday JS, Perrott MR, Symonds JE, Walker SP, Lovett B, Preece MA, Davie PS (2016) Unilateral perivertebral fibrosis associated with lordosis, kyphosis and scoliosis (LKS) in farmed Chinook salmon in New Zealand. Dis Aquat Organ 121:211–221. https://doi.org/10.3354/dao03056
Nassari S, Duprez D, Fournier-Thibault C (2017) Non-myogenic contribution to muscle development and homeostasis: the role of connective tissues. Front Cell Dev Biol 5. Article 22.https://doi.org/10.3389/fcell.2017.00022
Negatu Z, Meier AH (1995) In vitro incorporation of [14C] glycine into muscle protein of gulf killifish (Fundulus grandis) in response to insulin-like growth factor-I. Gen Comp Endocrinol 98:193–201. https://doi.org/10.1006/gcen.1995.1060
Olesen I, Gjedrem T, Bentsen HB, Gjerde B, Rye M (2003) Breeding programs for sustainable aquaculture. J Appl Aquacult 13:179–204. https://doi.org/10.1300/J028v13n03_01
Parsons DM, Sim-Smith CJ, Cryer M, Francis MP, Hartill B, Jones EG, Le Port A, Lowe M, McKenzie J, Morrison M, Paul LJ, Radford C, Ross PM, Spong KT, Trnski T, Usmar N, Walsh C, Zeldis J (2014) Snapper (Chrysophrys auratus): a review of life history and key vulnerabilities in New Zealand. N Z J Mar Freshwat Res 48:256–283. https://doi.org/10.1080/00288330.2014.892013
Paylor B, Natarajan A, Zhang R-H, Rossi F (2011) Chapter six - nonmyogenic cells in skeletal muscle regeneration. In Pavlath GK (ed) Current topics in developmental biology. Academic Press, pp 139–165. https://doi.org/10.1016/B978-0-12-385940-2.00006-1
Peng LM, Zheng Y, You F, Wu ZH, Tan X, Jiao S, Zhang PJ (2016) Comparison of growth characteristics between skeletal muscle satellite cell lines from diploid and triploid olive flounder Paralichthys olivaceus. PeerJ 4:e1519. https://doi.org/10.7717/peerj.1519
Potter G, Smith AST, Vo NTK, Muster J, Weston W, Bertero A, Maves L, Mack DL, Rostain A (2020) A more open approach is needed to develop cell-based fish technology: it starts with zebrafish. One Earth 3:54–64. https://doi.org/10.1016/j.oneear.2020.06.005
Rius-Francino M, Acerete L, Jiménez-Amilburu V, Capilla E, Navarro I, Gutiérrez J (2011) Differential effects on proliferation of GH and IGFs in sea bream (Sparus aurata) cultured myocytes. Gen Comp Endocrinol 172:44–49. https://doi.org/10.1016/j.ygcen.2011.03.024
Rougee L, Ostrander GK, Richmond RH, Lu Y (2007) Establishment, characterization, and viral susceptibility of two cell lines derived from goldfish Carassius auratus muscle and swim bladder. Dis Aquat Organ 77:127–135. https://doi.org/10.3354/dao01802
Rubenstein AB, Smith GR, Raue U, Begue G, Minchev K, Ruf-Zamojski F, Nair VD, Wang X, Zhou L, Zaslavsky E, Trappe TA, Trappe S, Sealfon SC (2020) Single-cell transcriptional profiles in human skeletal muscle. Sci Reps 10:229. https://doi.org/10.1038/s41598-019-57110-6
Rubio N, Datar I, Stachura D, Kaplan D, Krueger K (2019) Cell-based fish: a novel approach to seafood production and an opportunity for cellular agriculture. Front Sustain Food Sys 3:43. https://doi.org/10.3389/fsufs.2019.00043
Ruparelia AA, Ratnayake D, Currie PD (2020) Stem cells in skeletal muscle growth and regeneration in amniotes and teleosts: Emerging themes. WIREs Dev Biol 9:e365. https://doi.org/10.1002/wdev.365
Sandoval-Castillo J, Beheregaray LB, Wellenreuther M (2022) Genomic prediction of growth in a commercially, recreationally, and culturally important marine resource, the Australian snapper (Chrysophrys auratus). G3 (Bethesda) 12(3). https://doi.org/10.1093/g3journal/jkac015
Shefer G, Wleklinski-Lee M, Yablonka-Reuveni Z (2004) Skeletal muscle satellite cells can spontaneously enter, an alternative mesenchymal pathway. J Cell Sci 117:5393–5404. https://doi.org/10.1242/jcs.01419
Sultan SHA, Dyer C, Knight RD (2021) Notch signaling regulates muscle stem cell homeostasis and regeneration in a teleost fish. Front Cell Dev Biol 9:726281 https://doi.org/10.3389/fcell.2021.726281
Syverud BC, VanDusen KW, Larkin LM (2016) Growth factors for skeletal muscle tissue engineering. Cells Tissues Organs 202:169–179. https://doi.org/10.1159/000444671
Tsai C-C, Wu S-B, Kau H-C, Wei Y-H (2018) Essential role of connective tissue growth factor (CTGF) in transforming growth factor-β1 (TGF-β1)-induced myofibroblast transdifferentiation from Graves’ orbital fibroblasts. Sci Reps 8:7276. https://doi.org/10.1038/s41598-018-25370-3
Uezumi A, Ito T, Morikawa D, Shimizu N, Yoneda T, Segawa M, Yamaguchi M, Ogawa R, Matev MM, Miyagoe-Suzuki Y (2011) Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci 124:3654–3664. https://doi.org/10.1242/jcs.086629
Velez EJ, Lutfi E, Azizi S, Perello M, Salmeron C, Riera-Codina M, Ibarz A, Fernandez-Borras J, Blasco J, Capilla E, Navarro I, Gutierrez J (2017) Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 467:28–40. https://doi.org/10.1016/j.aquaculture.2016.07.004
Vélez EJ, Lutfi E, Jiménez-Amilburu V, Riera-Codina M, Capilla E, Navarro I, Gutiérrez J (2014) IGF-I and amino acids effects through TOR signaling on proliferation and differentiation of gilthead sea bream cultured myocytes. Gen Comp Endocrinol 205:296–304. https://doi.org/10.1016/j.ygcen.2014.05.024
Wang G, LaPatra S, Zeng L, Zhao Z, Lu Y (2003) Establishment, growth, cryopreservation and species of origin identification of three cell lines from white sturgeon, Acipenser transmontanus. Methods Cell Sci 25:211–220. https://doi.org/10.1007/s11022-004-9120-x
Wang L, Cao Z, Liu Y, **ang Y, Sun Y, Zhou Y, Wang S, Guo W (2020) Establishment and characterization of a new cell line from the muscle of humpback grouper (Cromileptes altivelis). Fish Physiol Biochem 46:1897–1907. https://doi.org/10.1007/s10695-020-00841-5
Wellenreuther M, Le Luyer J, Cook D, Ritchie PA, Bernatchez L (2019) Domestication and temperature modulate gene expression signatures and growth in the Australasian Snapper Chrysophrys auratus. G3 (Bethesda) 9:105–116. https://doi.org/10.1534/g3.118.200647
Wu Y, Zhang X, Salmon M, Lin X, Zehner ZE (2007) TGFbeta1 regulation of vimentin gene expression during differentiation of the C2C12 skeletal myogenic cell line requires Smads, AP-1 and Sp1 family members. Biochim Biophys Acta 1773:427–439. https://doi.org/10.1016/j.bbamcr.2006.11.017
Zhao Z, Lu Y (2006) Establishment and characterization of two cell lines from bluefin trevally Caranx melampygus. Dis Aquat Organ 68:91–100. https://doi.org/10.3354/dao068091
Zhao Z, Montgomery-Brock D, Lee CS, Lu Y (2003) Establishment, characterization and viral susceptibility of 3 new cell lines from snakehead, Channa striatus (Blooch). Methods Cell Sci 25:155–166. https://doi.org/10.1007/s11022-004-3804-0
Zimmerman AM, Lowery MS (1999) Hyperplastic development and hypertrophic growth of muscle fibers in the white seabass (Atractoscion nobilis). J Exp Zool 284:299–308. https://doi.org/10.1002/(sici)1097-010x(19990801)284:3%3c299::aid-jez7%3e3.0.co;2-6
Acknowledgements
We would like to acknowledge all the staff at PFR’s finfish facility who have assisted us during this research. We would also like to acknowledge Dr Michael Algie for constructive comments on the manuscript.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by a Royal Society of New Zealand Catalyst Seeding Fund and a Ministry of Business, Innovation and Employment Smart Idea (C11X1906) fund awarded to GD.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Editor: J Denry Sato
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chong, G.L.W., Böhmert, B., Lee, L.E.J. et al. A continuous myofibroblast precursor cell line from the tail muscle of Australasian snapper (Chrysophrys auratus) that responds to transforming growth factor beta and fibroblast growth factor. In Vitro Cell.Dev.Biol.-Animal 58, 922–935 (2022). https://doi.org/10.1007/s11626-022-00734-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-022-00734-2