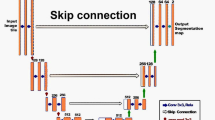
Abstract
Accurate liver and lesion segmentation plays a crucial role in the clinical assessment and therapeutic planning of hepatic diseases. The segmentation of the liver and lesions using automated techniques is a crucial undertaking that holds the potential to facilitate the early detection of malignancies and the effective management of patients’ treatment requirements by medical professionals. This research presents the Generalized U-Net (G-Unet), a unique hybrid model designed for segmentation tasks. The G-Unet model is capable of incorporating other models such as convolutional neural networks (CNN), residual networks (ResNets), and densely connected convolutional neural networks (DenseNet) into the general U-Net framework. The G-Unet model, which consists of three distinct configurations, was assessed using the LiTS dataset. The results indicate that G-Unet demonstrated a high level of accuracy in segmenting the data. Specifically, the G-Unet model, configured with DenseNet architecture, produced a liver tumor segmentation accuracy of 72.9% dice global score. This performance is comparable to the existing state-of-the-art methodologies. The study also showcases the influence of different preprocessing and postprocessing techniques on the accuracy of segmentation. The utilization of Hounsfield Unit (HU) windowing and histogram equalization as preprocessing approaches, together with the implementation of conditional random fields as postprocessing techniques, resulted in a notable enhancement of 3.35% in the accuracy of tumor segmentation.











Similar content being viewed by others
Data Availability
Dataset is freely available and can be downloaded from [41].
References
Almotairi S, Kareem G, Aouf M, Almutairi B, Salem MA-M (2020) Liver Tumor Segmentation in CT Scans Using Modified SegNet. Sensors (Basel) 20:1516. https://doi.org/10.3390/s20051516
Amin J, Anjum MA, Sharif M, Kadry S, Nadeem A, Ahmad SF (2022) Liver Tumor Localization Based on YOLOv3 and 3D-Semantic Segmentation Using Deep Neural Networks. Diagnostics 12:823. https://doi.org/10.3390/diagnostics12040823
Ayalew YA, Fante KA, Mohammed MA (2021) Modified U-Net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed Eng 3:4. https://doi.org/10.1186/s42490-021-00050-y
Bai Z, Jiang H, Li S, Yao Y-D (2019) Liver Tumor Segmentation Based on Multi-Scale Candidate Generation and Fractal Residual Network. IEEE Access 7:82122–82133. https://doi.org/10.1109/ACCESS.2019.2923218
Bi L, Kim J, Kumar A, Feng D (2017) Automatic liver lesion detection using cascaded deep residual networks. ar**v:1704.02703 [cs]
Budak Ü, Guo Y, Tanyildizi E, Şengür A (2020) Cascaded deep convolutional encoder-decoder neural networks for efficient liver tumor segmentation. Med Hypotheses 134:109431. https://doi.org/10.1016/j.mehy.2019.109431
Chen G, Li Z, Wang J, Wang J, Du S, Zhou J, Shi J, Zhou Y (2023) An improved 3D KiU-Net for segmentation of liver tumor. Comput Biol Med 160:107006. https://doi.org/10.1016/j.compbiomed.2023.107006
Chi J, Han X, Wu C, Wang H, Ji P (2021) X-Net: Multi-branch UNet-like network for liver and tumor segmentation from 3D abdominal CT scans. Neurocomputing 459:81–96. https://doi.org/10.1016/j.neucom.2021.06.021
Chlebus G, Meine H, Moltz JH, Schenk A (2017). Neural network-based automatic liver tumor segmentation with random forest-based candidate filtering. https://doi.org/10.48550/ar**v.1706.00842
Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Armbruster M, Hofmann F, D’Anastasi M, Sommer WH, Ahmadi S-A, Menze BH (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds) Medical Image computing and computer-assisted intervention – MICCAI 2016. Springer International Publishing, Cham, pp 415–423
Christ PF, Ettlinger F, Grün F, Elshaera MEA, Lipkova J, Schlecht S, Ahmaddy F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Hofmann F, Anastasi MD, Ahmadi S-A, Kaissis G, Holch J, Sommer W, Braren R, Heinemann V, Menze B (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. ar**v:1702.05970 [cs]
Di S, Zhao Y-Q, Liao M, Zhang F, Li X (2023) TD-Net: A Hybrid End-to-End Network for Automatic Liver Tumor Segmentation From CT Images. IEEE J Biomed Health Inform 27:1163–1172. https://doi.org/10.1109/JBHI.2022.3181974
Doggalli D, Sunil Kumar BS (2022) The Efficacy of U-Net in Segmenting Liver Tumors from Abdominal CT Images. IJIES 15:151–161. https://doi.org/10.22266/ijies2022.1031.14
Fan T, Wang G, Li Y, Wang H (2020) MA-Net: A Multi-Scale Attention Network for Liver and Tumor Segmentation. IEEE Access 8:179656–179665. https://doi.org/10.1109/ACCESS.2020.3025372
Han X (2017) Automatic Liver Lesion Segmentation Using A Deep Convolutional Neural Network Method. Med Phys 44:1408–1419. https://doi.org/10.1002/mp.12155
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR). pp 770–778
Jiang H, Shi T, Bai Z, Huang L (2019) AHCNet: an application of attention mechanism and hybrid connection for liver tumor segmentation in CT volumes. IEEE Access 7:24898–24909. https://doi.org/10.1109/ACCESS.2019.2899608
Jiang L, Ou J, Liu R, Zou Y, **e T, **ao H, Bai T (2023) RMAU-Net: Residual Multi-Scale Attention U-Net For liver and tumor segmentation in CT images. Comput Biol Med 158:106838. https://doi.org/10.1016/j.compbiomed.2023.106838
Kushnure DT, Talbar SN (2022) HFRU-Net: High-Level Feature Fusion and Recalibration UNet for Automatic Liver and Tumor Segmentation in CT Images. Comput Methods Programs Biomed 213:106501. https://doi.org/10.1016/j.cmpb.2021.106501
Li J, Liu K, Hu Y, Zhang H, Heidari AA, Chen H, Zhang W, Algarni AD, Elmannai H (2023) Eres-UNet++: Liver CT image segmentation based on high-efficiency channel attention and Res-UNet++. Comput Biol Med 158:106501. https://doi.org/10.1016/j.compbiomed.2022.106501
Li S, Tso GKF, He K (2020) Bottleneck feature supervised U-Net for pixel-wise liver and tumor segmentation. Expert Syst Appl 145:113131. https://doi.org/10.1016/j.eswa.2019.113131
Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng P-A (2018) H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging 37:2663–2674. https://doi.org/10.1109/TMI.2018.2845918
Li Y, Zou B, Liu Q (2021) A deep attention network via high-resolution representation for liver and liver tumor segmentation. Biocybern Biomed Eng 41:1518–1532. https://doi.org/10.1016/j.bbe.2021.08.010
Liu Y, Yang F, Yang Y (2023) A partial convolution generative adversarial network for lesion synthesis and enhanced liver tumor segmentation. J Appl Clin Med Phys 24:e13927. https://doi.org/10.1002/acm2.13927
Lv P, Wang J, Wang H (2022) 2.5D lightweight RIU-Net for automatic liver and tumor segmentation from CT. Biomed Signal Process Control 75:103567. https://doi.org/10.1016/j.bspc.2022.103567
Manjunath RV, Kwadiki K (2022) Modified U-NET on CT images for automatic segmentation of liver and its tumor. Biomed Eng Adv 4:100043. https://doi.org/10.1016/j.bea.2022.100043
Moghbel M, Mashohor S, Mahmud R, Saripan MIB (2018) Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev 50:497–537. https://doi.org/10.1007/s10462-017-9550-x
Rahman H, Bukht TFN, Imran A, Tariq J, Tu S, Alzahrani A (2022) A Deep Learning Approach for Liver and Tumor Segmentation in CT Images Using ResUNet. Bioengineering 9:368. https://doi.org/10.3390/bioengineering9080368
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. https://doi.org/10.48550/ARXIV.1505.04597
Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I (2022) Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol 77:1598–1606. https://doi.org/10.1016/j.jhep.2022.08.021
Seo H, Huang C, Bassenne M, **ao R, **ng L (2020) Modified U-Net (mU-Net) with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images. IEEE Trans Med Imaging 39:1316–1325. https://doi.org/10.1109/TMI.2019.2948320
Shu X, Yang Y, Liu J, Chang X, Wu B (2023) ALVLS: Adaptive local variances-Based levelset framework for medical images segmentation. Pattern Recognit 136:109257. https://doi.org/10.1016/j.patcog.2022.109257
Shu X, Yang Y, Wu B (2021) Adaptive segmentation model for liver CT images based on neural network and level set method. Neurocomputing 453:438–452. https://doi.org/10.1016/j.neucom.2021.01.081
Song L, Wang H, Wang ZJ (2021) Bridging the Gap Between 2D and 3D Contexts in CT Volume for Liver and Tumor Segmentation. IEEE J Biomed Health Inform 25:3450–3459. https://doi.org/10.1109/JBHI.2021.3075752
Sun C, Guo S, Zhang H, Li J, Chen M, Ma S, ** L, Liu X, Li X, Qian X (2017) Automatic segmentation of liver tumors from multiphase contrast-enhanced CT images based on FCNs. Artif Intell Med 83:58–66. https://doi.org/10.1016/j.artmed.2017.03.008
Tran S-T, Cheng C-H, Liu D-G (2021) A multiple Layer U-Net, U n -Net, for liver and liver tumor segmentation in CT. IEEE Access 9:3752–3764. https://doi.org/10.1109/ACCESS.2020.3047861
Zhang C, Hua Q, Chu Y, Wang P (2021) Liver tumor segmentation using 2.5D UV-Net with multi-scale convolution. Comput Biol Med 133:104424. https://doi.org/10.1016/j.compbiomed.2021.104424
Zhang C, Lu J, Hua Q, Li C, Wang P (2022) SAA-Net: U-shaped network with Scale-Axis-Attention for liver tumor segmentation. Biomed Signal Process Control 73:103460. https://doi.org/10.1016/j.bspc.2021.103460
Zhang Y, Jiang B, Wu J, Ji D, Liu Y, Chen Y, Wu EX, Tang X (2020) Deep Learning Initialized and Gradient Enhanced Level-Set Based Segmentation for Liver Tumor From CT Images. IEEE Access 8:76056–76068. https://doi.org/10.1109/ACCESS.2020.2988647
Zhu Y, Yu A, Rong H, Wang D, Song Y, Liu Z, Sheng VS (2021) Multi-Resolution Image Segmentation Based on a Cascaded U-ADenseNet for the Liver and Tumors. JPM 11:1044. https://doi.org/10.3390/jpm11101044
CodaLab - Competition. https://competitions.codalab.org/competitions/17094
Funding
No Funding was obtained for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflict of interests on the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
D J, D., B S, S.K. Liver tumor segmentation using G-Unet and the impact of preprocessing and postprocessing methods. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18759-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18759-y