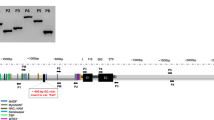
Abstract
Malt barley (Hordeum vulgare L.) is an important cash crop with stringent grain quality standards. Timing of the switch from vegetative to reproductive growth and timing of whole-plant senescence and nutrient remobilization are critical for cereal grain yield and quality. Understanding the genetic variation in genes associated with these developmental traits can streamline genotypic selection of superior malt barley germplasm. Here, we determined the effects of allelic variation in three genes encoding a glycine-rich RNA-binding protein (HvGR-RBP1) and two NAC transcription factors (HvNAM1 and HvNAM2) on malt barley agronomics and quality using previously developed markers for HvGR-RBP1 and HvNAM1 and a novel marker for HvNAM2. Based on a single-nucleotide polymorphism (SNP) in the first intron, the utilized marker differentiates NAM2 alleles of low-grain protein variety ‘Karl’ and of higher protein variety ‘Lewis’. We demonstrate that the selection of favorable alleles for each gene impacts heading date, senescence timing, grain size, grain protein concentration, and malt quality. Specifically, combining ‘Karl’ alleles for the two NAC genes with the ‘Lewis’ HvGR-RBP1 allele extends grain fill duration, increases the percentage of plump kernels, decreases grain protein, and provides malt quality stability. Molecular markers for these genes are therefore highly useful tools in malt barley breeding.

Similar content being viewed by others
Data availability
HvNAM2 sequences from varieties ‘Karl’ and ‘Lewis’ are shown in Online Resource 6. Genoty** data for HvGR-RBP1, HvNAM1, and HvNAM2 are shown in Online Resource 1; agronomic and malt quality data for experiment 1 (Pauli et al. 2015) are shown in Online Resources 2 and 4, and agronomic data for experiment 2 (Alptekin et al. 2021) are shown in Online Resource 3.
Code availability
No new code was generated as a part of this study.
References
Alptekin B, Mangel D, Pauli D, Blake T, Lachowiec J, Hoogland T, Fischer A, Sherman J (2021) Combined effects of a glycine-rich RNA-binding protein and a NAC transcription factor extend grain fill duration and improve malt barley agronomic performance. Theor Appl Genet 134:351–366. https://doi.org/10.1007/s00122-020-03701-1
Ayoub M, Symons J, Edney J, Mather E (2002) QTLs affecting kernel size and shape in a two-rowed by six-rowed barley cross. Theor Appl Genet 105:237–247. https://doi.org/10.1007/s00122-002-0941-1
Baik B-K, Ullrich SE (2008) Barley for food: characteristics, improvement, and renewed interest. J Cereal Sci 48:233–242. https://doi.org/10.1016/j.jcs.2008.02.002
Bingham IJ, Blake J, Foulkes MJ, Spink J (2007) Is barley yield in the UK sink limited? II. Factors affecting potential grain size. Field Crops Res 101:212–220. https://doi.org/10.1016/j.fcr.2006.11.004
Brouwer BO, Schwarz PB, Barr JM, Hayes PM, Murphy KM, Jones SS (2016) Evaluating barley for the emerging craft malting industry in western Washington. Agron J 108:939–949. https://doi.org/10.2134/agronj2015.0385
Burger WC, Wesenberg DM, Carden JE, Pawlisch PE (1979) Protein content and composition of Karl and related barleys. Crop Sci 19:235–238. https://doi.org/10.2135/cropsci1979.0011183X001900020016x
Cai S, Yu G, Chen X, Huang Y, Jiang X, Zhang G, ** X (2013) Grain protein content variation and its association analysis in barley. BMC Plant Biol 13:35. https://doi.org/10.1186/1471-2229-13-35
Cao S, Jiang L, Song S, **g R, Xu G (2006) AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis Cell Mol Biol Lett 11:526–535. https://doi.org/10.2478/s11658-006-0042-2
Chappell A, Scott KP, Griffiths IA, Cowan AA, Hawes C, Wishart J, Martin P (2017) The agronomic performance and nutritional content of oat and barley varieties grown in a northern maritime environment depends on variety and growing conditions. J Cereal Sci 74:1–10. https://doi.org/10.1016/j.jcs.2017.01.005
Ciuzan O, Hancock J, Pamfil D, Wilson I, Ladomery M (2015) The evolutionarily conserved multifunctional glycine-rich RNA-binding proteins play key roles in development and stress adaptation. Physiol Plant 153:1–11. https://doi.org/10.1111/ppl.12286
Coventry SJ, Barr AR, Eglinton JK, McDonald GK (2003) The determinants and genome locations influencing grain weight and size in barley (Hordeum vulgare L.). Aust J Agric Res 54:1103–1115. https://doi.org/10.1071/ar02194
Daba S, Horsley R, Schwarz P, Chao S, Capettini F, Mohammadi M (2019) Association and genome analyses to propose putative candidate genes for malt quality traits. J Sci Food Agric 99:2775–2785. https://doi.org/10.1002/jsfa.9485
Delignette-Muller ML, Dutang C (2015) fitdistrplus: an R package for fitting distributions. J Stat Softw 64:1–34. https://doi.org/10.18637/jss.v064.i04
Distelfeld A, Avni R, Fischer AM (2014) Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot 65:3783–3798. https://doi.org/10.1093/jxb/ert477
Distelfeld A, Korol A, Dubcovsky J, Uauy C, Blake T, Fahima T (2008) Colinearity between the barley grain protein content (GPC) QTL on chromosome arm 6HS and the wheat Gpc-B1 region. Mol Breed 22:25–38. https://doi.org/10.1007/s11032-007-9153-3
Eagles HA, Bedggood AG, Panozzo JF, Martin PJ (1995) Cultivar and environmental effects on malting quality in barley. Aust J Agric Res 46:831–844. https://doi.org/10.1071/ar9950831
Elía M, Swanston JS, Moralejo M, Casas A, Pérez-Vendrell AM, Ciudad FJ, Thomas WTB, Smith PL, Ullrich SE, Molina-Cano JL (2010) A model of the genetic differences in malting quality between European and North American barley cultivars based on a QTL study of the cross Triumph × Morex. Plant Breed 129:280–290. https://doi.org/10.1111/j.1439-0523.2009.01694.x
Fan C, Zhai H, Wang H, Yue Y, Zhang M, Li J, Wen S, Guo G, Zeng Y, Ni Z, You M (2017) Identification of QTLs controlling grain protein concentration using a high-density SNP and SSR linkage map in barley (Hordeum vulgare L.). BMC Plant Biol 17:122. https://doi.org/10.1186/s12870-017-1067-6
Fang Y, Zhang X, Xue D (2019) Genetic analysis and molecular breeding applications of malting quality QTLs in barley. Front Genet 10:352. https://doi.org/10.3389/fgene.2019.00352
Freytes SN, Canelo M, Cerdán PD (2021) Regulation of flowering time: when and where? Curr Opin Plant Biol 63:102049. https://doi.org/10.1016/j.pbi.2021.102049
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612. https://doi.org/10.1111/j.1365-313X.2006.02723.x
Gupta M, Abu-Ghannam N, Gallaghar E (2010) Barley for brewing: characteristic changes during malting, brewing and applications of its by-products. Compr Rev Food Sci Food Saf 9:318–328. https://doi.org/10.1111/j.1541-4337.2010.00112.x
Hagenblad J, Vanhala T, Madhavan S, Leino MW (2022) Protein content and HvNAM alleles in Nordic barley (Hordeum vulgare) during a century of breeding. Hereditas 159:12. https://doi.org/10.1186/s41065-022-00227-y
Hill CB, Li C (2016) Genetic architecture of flowering phenology in cereals and opportunities for crop improvement. Front Plant Sci 7:1906. https://doi.org/10.3389/fpls.2016.01906
Hockett EA, Gilbertson KM, McGuire CF, Bergman LE, Wiesner LE, Robbins GS (1985) Registration of ‘Lewis’ barley. Crop Sci 25:570–571. https://doi.org/10.2135/cropsci1985.0011183X002500030036x
Homan MM (2004) Beer and its drinkers: an ancient Near Eastern love story. Near East Archaeol 67:84–95. https://doi.org/10.2307/4132364
Jensen MK, Skriver K (2014) NAC transcription factor gene regulatory and protein-protein interaction networks in plant stress responses and senescence. IUBMB Life 66:156–166. https://doi.org/10.1002/iub.1256
Jukanti AK, Fischer AM (2008) A high-grain protein content locus on barley (Hordeum vulgare) chromosome 6 is associated with increased flag leaf proteolysis and nitrogen remobilization. Physiol Plant 132:426–439. https://doi.org/10.1111/j.1399-3054.2007.01044.x
Jukanti AK, Heidlebaugh NM, Parrott DL, Fischer IA, McInnerney K, Fischer AM (2008) Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol 177:333–349. https://doi.org/10.1111/j.1469-8137.2007.02270.x
Kim JS, Jung HJ, Lee HJ, Kim KA, Goh C-H, Woo Y, Oh SH, Han YS, Kang H (2008) Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana Plant J 55:455–466. https://doi.org/10.1111/j.1365-313X.2008.03518.x
Lacerenza JA, Parrott DL, Fischer AM (2010) A major grain protein content locus on barley (Hordeum vulgare L.) chromosome 6 influences flowering time and sequential leaf senescence. J Exp Bot 61:3137–3149. https://doi.org/10.1093/jxb/erq139
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. https://doi.org/10.1093/nar/gkz268
Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, ** D, Chen X, Ming F (2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174:1747–1763. https://doi.org/10.1104/pp.17.00542
Mascher M, Gundlach H, Himmelbach A et al (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544:427–433. https://doi.org/10.1038/nature22043
Mickelson S, See D, Meyer FD, Garner JP, Foster CR, Blake TK, Fischer AM (2003) Map** of QTL associated with nitrogen storage and remobilization in barley (Hordeum vulgare L.) leaves. J Exp Bot 54:801–812. https://doi.org/10.1093/jxb/erg084
Nice L, Huang Y, Steffenson BJ, Gyenis L, Schwarz P, Smith KP, Muehlbauer GJ (2019) Map** malting quality and yield characteristics in a north American two-rowed malting barley × wild barley advanced backcross population. Mol Breed 39:121. https://doi.org/10.1007/s11032-019-1030-3
O’Donovan JT, Clayton GW, Grant CA, Harker KN, Turkington TK, Lupwayi NZ (2008) Effect of nitrogen rate and placement and seeding rate on barley productivity and wild oat fecundity in a zero tillage system. Crop Sci 48:1569–1574. https://doi.org/10.2135/cropsci2007.10.0587
Ochoa A, Storey JD (2021) Estimating FST and kinship for arbitrary population structures. PLoS Genet 17:e1009241. https://doi.org/10.1371/journal.pgen.1009241
Parrott DL, Downs EP, Fischer AM (2012) Control of barley (Hordeum vulgare L.) development and senescence by the interaction between a chromosome six grain protein content locus, day length, and vernalization. J Exp Bot 63:1329–1339. https://doi.org/10.1093/jxb/err360
Pauli D, Brown-Guedira G, Blake TK (2015) Identification of malting quality QTLs in advanced generation breeding germplasm. J Am Soc Brew Chem 73:29–40. https://doi.org/10.1094/asbcj-2015-0129-01
Peterson RA, Cavanaugh JE (2020) Ordered quantile normalization: a semiparametric transformation built for the cross-validation era. J Appl Stat 47:2312–2327. https://doi.org/10.1080/02664763.2019.1630372
Podzimska-Sroka D, O’Shea C, Gregersen P, Skriver K (2015) NAC transcription factors in senescence: from molecular structure to function in crops. Plants 4:412–448. https://doi.org/10.3390/plants4030412
Sakuraba Y, Piao W, Lim J-H, Han S-H, Kim Y-S, An G, Paek N-C (2015) Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol 56:2325–2339. https://doi.org/10.1093/pcp/pcv144
Schüttpelz M, Schöning JC, Doose S, Neuweiler H, Peters E, Staiger D, Sauer M (2008) Changes in conformational dynamics of mRNA upon AtGRP7 binding studied by fluorescence correlation spectroscopy. J Am Chem Soc 130:9507–9513. https://doi.org/10.1021/ja801994z
See D, Kanazin V, Kephart K, Blake T (2002) Map** genes controlling variation in barley grain protein concentration. Crop Sci 42:680–685. https://doi.org/10.2135/cropsci2002.6800
Steffen A, Elgner M, Staiger D (2019) Regulation of flowering time by the RNA-binding proteins AtGRP7 and AtGRP8. Plant Cell Physiol 60:2040–2050. https://doi.org/10.1093/pcp/pcz124
Stevens WB, Sainju UM, Caesar-TonThat T, Iversen WM (2015) Malt barley yield and quality affected by irrigation, tillage, crop rotation, and nitrogen fertilization. Agron J 107:2107–2119. https://doi.org/10.2134/agronj15.0027
Strader L, Weijers D, Wagner D (2022) Plant transcription factors - being in the right place with the right company. Curr Opin Plant Biol 65:102136. https://doi.org/10.1016/j.pbi.2021.102136
Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D (2008) The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana Plant J 56:239–250. https://doi.org/10.1111/j.1365-313X.2008.03591.x
Tripet BP, Mason KE, Eilers BJ, Burns J, Powell P, Fischer AM, Copié V (2014) Structural and biochemical analysis of the Hordeum vulgare L. HvGR-RBP1 protein, a glycine-rich RNA-binding protein involved in the regulation of barley plant development and stress response. Biochemistry 53:7945–7960. https://doi.org/10.1021/bi5007223
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301. https://doi.org/10.1126/science.1133649
Walker CK, Ford R, Muñoz-Amatriain M, Panozzo JF (2013) The detection of QTLs in barley associated with endosperm hardness, grain density, grain size and malting quality using rapid phenoty** tools. Theor Appl Genet 126:2533–2551. https://doi.org/10.1007/s00122-013-2153-2
Waters BM, Uauy C, Dubcovsky J, Grusak MA (2009) Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J Exp Bot 60:4263–4274. https://doi.org/10.1093/jxb/erp257
Wesenberg DM, Hayes RM, Standridge NN, Burger WC, Goplin ED, Petr FC (1976) Registration of Karl barley (Reg. No. 147). Crop Sci 16:737. https://doi.org/10.2135/cropsci1976.0011183X001600050039x
Weston DT, Horsley RD, Schwarz PB, Goos RJ (1993) Nitrogen and planting date effects on low-protein spring barley. Agron J 85:1170–1174. https://doi.org/10.2134/agronj1993.00021962008500060015x
Wimmer V, Albrecht T, Auinger H-J, Schön C-C (2012) synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics 28:2086–2087. https://doi.org/10.1093/bioinformatics/bts335
Woods DP, Amasino R (2015) Dissecting the control of flowering time in grasses using Brachypodium distachyon. In: Vogel J (ed) Genetics and genomics of Brachypodium. Springer, Cham, pp 259–273. https://doi.org/10.1007/7397_2015_10
Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, Fernie AR, Kaufmann K, Xue GP, Mueller-Roeber B, Balazadeh S (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis Plant Cell 24:482–506. https://doi.org/10.1105/tpc.111.090894
Yang DH, Kwak KJ, Kim MK, Park SJ, Yang KY, Kang H (2014) Expression of Arabidopsis glycine-rich RNA-binding protein AtGRP2 or AtGRP7 improves grain yield of rice (Oryza sativa) under drought stress conditions. Plant Sci 214:106–112. https://doi.org/10.1016/j.plantsci.2013.10.006
Ziyatdinov A, Vázquez-Santiago M, Brunel H, Martinez-Perez A, Aschard H, Soria JM (2018) lme4qtl: linear mixed models with flexible covariance structure for genetic studies of related individuals. BMC Bioinform 19:68. https://doi.org/10.1186/s12859-018-2057-x
Acknowledgements
The authors would like to thank Christopher Martens for malt phenoty**.
Funding
This project was supported by funding from the Montana Agricultural Experiment Station (MAES), the Montana Wheat and Barley Committee, the Brewers Association, the American Malting Barley Association, and the United States of Agriculture—National Institute of Food and Agriculture (USDA-NIFA) (2017–68008-26209).
Author information
Authors and Affiliations
Contributions
BA contributed to HvNAM2 sequencing and marker development, performed HvNAM1 and HvGR-RBP1 genoty**, performed all statistical analyses (guided and supported by JL), and wrote a draft manuscript. ME contributed to HvNAM2 sequencing, developed the VNTR marker, and performed HvNAM2 genoty**. DM performed experiment 2 field studies, supported by JS’s breeding program. DP and TB shared field data for experiment 1. HT helped with malt phenoty**. AMF and JS obtained funding and directed the study. All authors have reviewed and approved the manuscript, as submitted to Molecular Breeding.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alptekin, B., Erfatpour, M., Mangel, D. et al. Selection of favorable alleles of genes controlling flowering and senescence improves malt barley quality. Mol Breeding 42, 59 (2022). https://doi.org/10.1007/s11032-022-01331-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-022-01331-7