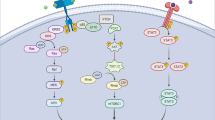
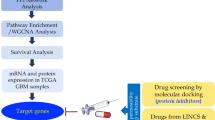
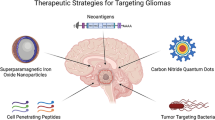
Abstract
Amongst the several types of brain cancers known to humankind, glioma is one of the most severe and life-threatening types of cancer, comprising 40% of all primary brain tumors. Recent reports have shown the incident rate of gliomas to be 6 per 100,000 individuals per year globally. Despite the various therapeutics used in the treatment of glioma, patient survival rate remains at a median of 15 months after undergoing first-line treatment including surgery, radiation, and chemotherapy with Temozolomide. As such, the discovery of newer and more effective therapeutic agents is imperative for patient survival rate. The advent of computer-aided drug design in the development of drug discovery has emerged as a powerful means to ascertain potential hit compounds with distinctively high therapeutic effectiveness against glioma. This review encompasses the recent advances of bio-computational in-silico modeling that have elicited the discovery of small molecule inhibitors and/or drugs against various therapeutic targets in glioma. The relevant information provided in this report will assist researchers, especially in the drug design domains, to develop more effective therapeutics against this global disease.


Similar content being viewed by others
Data Availability
Data supporting this review can be found in the list of references.
Code Availability
Not Applicable.
References
Liang J, Lv X, Lu C et al (2020) Prognostic factors of patients with Gliomas—an analysis on 335 patients with Glioblastoma and other forms of Gliomas. BMC Cancer 20:1–7. https://doi.org/10.1186/s12885-019-6511-6
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Boele FW, Klein M, Reijneveld JC et al (2014) Symptom management and quality of life in glioma patients. CNS Oncol 3:37–47
Weller M, van den Bent M, Preusser M et al (2021) EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 18:170–186. https://doi.org/10.1038/s41571-020-00447-z
Goldbrunner R, Ruge M, Kocher M et al (2018) The treatment of gliomas in adulthood. Dtsch Arztebl Int. https://doi.org/10.3238/arztebl.2018.0356
Holland EC (2001) Progenitor cells and glioma formation. Curr Opin Neurol 14:683–688. https://doi.org/10.1097/00019052-200112000-00002
Louis DN, Ohgaki H, Wiestler OD et al (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114:97–109. https://doi.org/10.1007/s00401-007-0243-4
Zong H, Verhaak RGW, Canolk P (2012) The cellular origin for malignant glioma and prospects for clinical advancements. Expert Rev Mol Diagn 12:383–394. https://doi.org/10.1586/erm.12.30
Weller M, Wick W, Aldape K et al (2015) Glioma. Nat Rev Dis Prim. https://doi.org/10.1038/nrdp.2015.17
Fiorentini A, Capelli D, Saraceni F et al (2020) The time has come for targeted therapies for AML: lights and shadows. Oncol Ther 8:13–32. https://doi.org/10.1007/s40487-019-00108-x
Placone AL, Quiñones-Hinojosa A, Searson PC (2016) The role of astrocytes in the progression of brain cancer: complicating the picture of the tumor microenvironment. Tumor Biol 37:61–69. https://doi.org/10.1007/s13277-015-4242-0
Wei J, Gabrusiewicz K, Heimberger A (2013) The controversial role of microglia in malignant gliomas. Clin Dev Immunol. https://doi.org/10.1155/2013/285246
Li W, Graeber MB (2012) The molecular profile of microglia under the influence of glioma. Neuro Oncol 14:958–978. https://doi.org/10.1093/neuonc/nos116
Rathke-Hartlieb S, Budde P, Ewert S et al (2000) Elevated expression of membrane type 1 metalloproteinase (MT1-MMP) in reactive astrocytes following neurodegeneration in mouse central nervous system. FEBS Lett 481:227–234. https://doi.org/10.1016/S0014-5793(00)02011-1
Coniglio SJ, Segall JE (2013) Review: molecular mechanism of microglia stimulated glioblastoma invasion. Matrix Biol 32:372–380. https://doi.org/10.1016/j.matbio.2013.07.008
Louis DN (2006) Molecular pathology of malignant gliomas. Annu Rev Pathol 1:97–117. https://doi.org/10.1146/annurev.pathol.1.110304.100043
Furnari FB, Fenton T, Bachoo RM et al (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21:2683–2710. https://doi.org/10.1101/gad.1596707
Reifenberger G, Collins VP (2004) Pathology and molecular genetics of astrocytic gliomas. J Mol Med 82:656–670. https://doi.org/10.1007/s00109-004-0564-x
Gupta A, Dwivedi T (2017) A simplified overview of WHO classification update. J Neurosci Rural Pract 8:4103. https://doi.org/10.4103/jnrp.jnrp
** Y, **ao W, Song T et al (2016) Expression and prognostic significance of p53 in glioma patients: a meta-analysis. Neurochem Res 41:1723–1731. https://doi.org/10.1007/s11064-016-1888-y
Hartmann C, Mueller W, Von Deimling A (2004) Pathology and molecular genetics of oligodendroglial tumors. J Mol Med 82:638–655. https://doi.org/10.1007/s00109-004-0565-9
Holleczek B, Zampella D, Urbschat S et al (2019) Incidence, mortality and outcome of meningiomas: a population-based study from Germany. Cancer Epidemiol. https://doi.org/10.1016/j.canep.2019.07.001
Simpson D (1957) The recurrence of intracranial meningiomas after surgical treatment. J Neurol Neurosurg Psychiatry 20:22–39. https://doi.org/10.1136/jnnp.20.1.22
Aflorei ED, Korbonits M (2014) Epidemiology and etiopathogenesis of pituitary adenomas. J Neurooncol 117:379–394. https://doi.org/10.1007/s11060-013-1354-5
Varlamov EV, McCartney S, Fleseriu M (2019) Functioning pituitary adenomas—current treatment options and emerging medical therapies. Eur Endocrinol 15:30–40. https://doi.org/10.17925/EE.2019.15.1.30
Fakhran S, Escott EJ (2008) Pineocytoma mimicking a pineal cyst on imaging: true diagnostic dilemma or a case of incomplete imaging? Am J Neuroradiol 29:159–163. https://doi.org/10.3174/ajnr.A0750
Orr BA (2020) Pathology, diagnostics, and classification of medulloblastoma. Brain Pathol 30:664–678. https://doi.org/10.1111/bpa.12837
Franceschi E, Seidel C, Sahm F et al (2021) How we treat medulloblastoma in adults. ESMO Open 6:100173. https://doi.org/10.1016/j.esmoop.2021.100173
Bagley CA, Connors SW, Aoun SG et al (2020) Recent advances in understanding and managing chordomas: an update. F1000Research 9:1–7. https://doi.org/10.12688/f1000research.22440.1
Heery CR (2016) Chordoma: the quest for better treatment options. Oncol Ther 4:35–51. https://doi.org/10.1007/s40487-016-0016-0
Bakker SH, Jacobs WCH, Pondaag W et al (2018) Chordoma: a systematic review of the epidemiology and clinical prognostic factors predicting progression-free and overall survival. Eur Spine J 27:3043–3058. https://doi.org/10.1007/s00586-018-5764-0
Cervio A, Villalonga J, Mormandi R et al (2017) Surgical treatment of cerebellar hemangioblastomas. Surg Neurol Int. https://doi.org/10.4103/sni.sni_490_16
Vigneswaran K, Neill S, Hadjipanayis CG (2015) Beyond the World Health Organization grading of infiltrating gliomas: advances in the molecular genetics of glioma classification. Ann Transl Med 3:95. https://doi.org/10.3978/j.issn.2305-5839.2015.03.57
Zhu Y, Liu X, Yang P et al (2014) Alkylglyceronephosphate synthase (AGPS) alters lipid signaling pathways and supports chemotherapy resistance of glioma and hepatic carcinoma cell lines. Asian Pac J Cancer Prev 15:3219–3226
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Gilbert MR, Ruda R, Soffietti R (2010) Ependymomas in adults. Curr Neurol Neurosci Rep 10:240–247. https://doi.org/10.1007/s11910-010-0109-3
Wen PY, Huse JT (2017) 2016 World Health Organization classification of central nervous system tumors. Contin Lifelong Learn Neurol 23:1531–1547. https://doi.org/10.1212/CON.0000000000000536
Yu W, Zhang L, Wei Q, Shao A (2020) O6-methylguanine-DNA methyltransferase (MGMT): challenges and new opportunities in glioma chemotherapy. Front Oncol 9:1–11. https://doi.org/10.3389/fonc.2019.01547
Zhang P, **a Q, Liu L et al (2020) Current opinion on molecular characterization for GBM classification in guiding clinical diagnosis, prognosis, and therapy. Front Mol Biosci 7:1–13. https://doi.org/10.3389/fmolb.2020.562798
Ward PS, Patel J, Wise DR et al (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17:225–234. https://doi.org/10.1016/j.ccr.2010.01.020
Aquilanti E, Miller J, Santagata S et al (2018) Updates in prognostic markers for gliomas. Neuro Oncol 20:VII17–VII26. https://doi.org/10.1093/neuonc/noy158
Ludwig K, Kornblum HI (2017) Molecular markers in glioma. J Neurooncol 134:505–512. https://doi.org/10.1007/s11060-017-2379-y
Nandakumar P, Mansouri A, Das S (2017) The role of ATRX in glioma biology. Front Oncol 7:1–8. https://doi.org/10.3389/fonc.2017.00236
Nieder C, Petersen S, Petersen C, Thames HD (2000) The challenge of p53 as prognostic and predictive factor in gliomas. Cancer Treat Rev 26:67–73. https://doi.org/10.1053/ctrv.1999.0145
McNamara MG, Sahebjam S, Mason WP (2013) Emerging biomarkers in glioblastoma. Cancers (Basel) 5:1103–1119. https://doi.org/10.3390/cancers5031103
Hung KS, Hong CY, Lee J et al (2000) Expression of p16(INK4A) induces dominant suppression of glioblastoma growth in situ through necrosis and cell cycle arrest. Biochem Biophys Res Commun 269:718–725. https://doi.org/10.1006/bbrc.2000.2339
Vuong HG, Altibi AMA, Duong UNP et al (2018) BRAF mutation is associated with an improved survival in glioma—a systematic review and meta-analysis. Mol Neurobiol 55:3718–3724. https://doi.org/10.1007/s12035-017-0599-y
Chowdhary M, Ene C, Silbergeld D (2015) Treatment of Gliomas: how did we get here? Surg Neurol Int 6:S85–S88. https://doi.org/10.4103/2152-7806.151348
Wang HY, Tang K, Liang TY et al (2016) The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J Exp Clin Cancer Res 35:1–9. https://doi.org/10.1186/s13046-016-0362-7
Vollmann-Zwerenz A, Leidgens V, Feliciello G et al (2020) Tumor cell invasion in glioblastoma. Int J Mol Sci 21:1–21. https://doi.org/10.3390/ijms21061932
Nanegrungsunk D, Onchan W, Chattipakorn N, Chattipakorn SC (2015) Current evidence of temozolomide and bevacizumab in treatment of gliomas. Neurol Res 37:167–183. https://doi.org/10.1179/1743132814Y.0000000423
Patterson J, Wongsurawat T, Rodriguez A (2020) A glioblastoma genomics primer for clinicians. Med Res Arch 8:1–13. https://doi.org/10.18103/mra.v8i2.2034
Dhermain F, Barani IJ (2016) Complications from radiotherapy. Handb Clin Neurol 134:219–234. https://doi.org/10.1016/B978-0-12-802997-8.00013-X
Belter A, Barciszewski J, Barciszewska AM (2020) Revealing the epigenetic effect of temozolomide on glioblastoma cell lines in therapeutic conditions. PLoS ONE 15:6–8. https://doi.org/10.1371/journal.pone.0229534
Zhang J, Stevens MFG, Bradshaw DT (2012) Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol 5:102–114. https://doi.org/10.2174/1874-470211205010102
World C (2009) Temozolomide birth of a blockbuster. Chem World 6:48–51
Newlands ES, Stevens MFG, Wedge SR et al (1997) Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev 23:35–61. https://doi.org/10.1016/S0305-7372(97)90019-0
Hanif F, Muzaffar K, Perveen K et al (2017) Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev 18:3–9. https://doi.org/10.22034/APJCP.2017.18.1.3
Stevens MFG, Hickman JA, Stone R et al (1984) Antitumor Imidazotetrazines. 1. Synthesis and chemistry of 8-carbamoyl-3-(2-chloroethyl)imidazo[5, l-d]-l,2,3,5-tetrazin-4(3H)-one, a novel broad-spectrum antitumor agent. J Med Chem 27:196–201. https://doi.org/10.1021/jm00368a016
Zhu P, Du XL, Lu G, Zhu JJ (2017) Survival benefit of glioblastoma patients after FDA approval of temozolomide concomitant with radiation and bevacizumab: a population-based study. Oncotarget 8:44015–44031. https://doi.org/10.18632/oncotarget.17054
Hottinger AF, Abdullah KG, Stupp R (2016) Current standards of care in glioblastoma therapy. Elsevier Inc, Amsterdam
Jiapaer S, Furuta T, Tanaka S et al (2018) Potential strategies overcoming the temozolomide resistance for glioblastoma. Neurol Med Chir (Tokyo) 58:405–421. https://doi.org/10.2176/nmc.ra.2018-0141
Wang D, Wang C, Wang L, Chen Y (2019) A comprehensive review in improving delivery of small-molecule chemotherapeutic agents overcoming the blood-brain/brain tumor barriers for glioblastoma treatment. Drug Deliv 26:551–565. https://doi.org/10.1080/10717544.2019.1616235
Raisa N, Marhaendraputro EA (2019) The side effects of chemotherapy in glioma. MNJ Malang Neurol J 5:92–97. https://doi.org/10.21776/ub.mnj.2019.005.02.9
Tseng W-L, Hsu H-H, Chen Y, Tseng S-H (2017) Tumor recurrence in a glioblastoma patient after discontinuation of prolonged temozolomide treatment. Asian J Neurosurg 12:727. https://doi.org/10.4103/ajns.ajns_39_15
Jain KK (2018) A critical overview of targeted therapies for glioblastoma. Front Oncol 8:1–19. https://doi.org/10.3389/fonc.2018.00419
Cohen MH, Shen YL, Keegan P, Pazdur R (2009) FDA Drug approval summary: bevacizumab (Avastin®) as treatment of recurrent glioblastoma multiforme. Oncologist 14:1131–1138. https://doi.org/10.1634/theoncologist.2009-0121
Kim MM, Umemura Y, Leung D (2018) Bevacizumab and glioblastoma past, present, and future directions. Cancer J (United States) 24:180–186. https://doi.org/10.1097/ppo.0000000000000326
Huang B, Li X, Li Y et al (2021) Current immunotherapies for glioblastoma multiforme. Front Immunol 11:1–12. https://doi.org/10.3389/fimmu.2020.603911
Romani M, Pistillo MP, Carosio R et al (2018) Immune checkpoints and innovative therapies in glioblastoma. Front Oncol 8:1–8. https://doi.org/10.3389/fonc.2018.00464
Kanu OO, Mehta A, Di C et al (2009) Glioblastoma multiforme: a review of therapeutic targets. Expert Opin Ther Targets 13:701–718. https://doi.org/10.1517/14728220902942348
Robles Irizarry L, Hambardzumyan D, Nakano I et al (2012) Therapeutic targeting of VEGF in the treatment of glioblastoma. Expert Opin Ther Targets 16:973–984. https://doi.org/10.1517/14728222.2012.711817
Jimenez-Pascual A, Siebzehnrubl F (2019) Fibroblast growth factor receptor functions in glioblastoma. Cells 8:715. https://doi.org/10.3390/cells8070715
Navis AC, Van Den Eijnden M, Schepens JTG et al (2010) Protein tyrosine phosphatases in glioma biology. Acta Neuropathol 119:157–175. https://doi.org/10.1007/s00401-009-0614-0
Chen L, Zhang W, He L et al (2020) Effect of alkylglycerone phosphate synthase on the expression levels of lncRNAs in glioma cells and its functional prediction. Oncol Lett 20:300211. https://doi.org/10.3892/ol.2020.11927
Benjamin DI, Cozzo A, Ji X et al (2013) Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc Natl Acad Sci USA 110:14912–14917. https://doi.org/10.1073/pnas.1310894110
Konteatis Z, Artin E, Nicolay B et al (2020) Vorasidenib (AG-881): a first-in-class, brain-penetrant dual inhibitor of mutant IDH1 and 2 for treatment of glioma. ACS Med Chem Lett 11:101–107. https://doi.org/10.1021/acsmedchemlett.9b00509
Padiadpu J, Mishra M, Sharma E et al (2016) Probing the druggability limits for enzymes of the NAD biosynthetic network in glioma. J Chem Inf Model 56:843–853. https://doi.org/10.1021/acs.jcim.5b00733
Franceschi E, Cavallo G, Lonardi S et al (2007) Gefitinib in patients with progressive high-grade gliomas: a multicentre phase II study by Gruppo Italiano Cooperativo di Neuro-Oncologia (GICNO). Br J Cancer 96:1047–1051. https://doi.org/10.1038/sj.bjc.6603669
Mendelsohn J, Baselga J (2003) Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol 21:2787–2799. https://doi.org/10.1200/JCO.2003.01.504
Makhlin I, Salinas RD, Zhang D et al (2019) Clinical activity of the EGFR tyrosine kinase inhibitor osimertinib in EGFR-mutant glioblastoma. CNS Oncol 8:CNS43. https://doi.org/10.2217/cns-2019-0014
Liu X, Chen X, Shi L et al (2019) The third-generation EGFR inhibitor AZD9291 overcomes primary resistance by continuously blocking ERK signaling in glioblastoma. J Exp Clin Cancer Res 38:1–14. https://doi.org/10.1186/s13046-019-1235-7
Endersby R, Whitehouse J, Hii H et al (2018) A pre-clinical assessment of the Pan-ERBB inhibitor dacomitinib in pediatric and adult brain tumors. Neoplasia (United States) 20:432–442. https://doi.org/10.1016/j.neo.2018.02.004
Sepúlveda JM, Sánchez-Gómez P, Vaz Salgado MÁ et al (2018) Dacomitinib: an investigational drug for the treatment of glioblastoma. Expert Opin Investig Drugs 27:823–829. https://doi.org/10.1080/13543784.2018.1528225
Vengoji R, Macha MA, Nimmakayala RK et al (2019) Afatinib and Temozolomide combination inhibits tumorigenesis by targeting EGFRvIII-cMet signaling in glioblastoma cells. J Exp Clin Cancer Res 38:1–13. https://doi.org/10.1186/s13046-019-1264-2
Shen J, Zhang T, Cheng Z et al (2018) Lycorine inhibits glioblastoma multiforme growth through EGFR suppression. J Exp Clin Cancer Res 37:1–19. https://doi.org/10.1186/s13046-018-0785-4
Gerstner ER, Eichler AF, Plotkin SR et al (2011) Phase I trial with biomarker studies of vatalanib (PTK787) in patients with newly diagnosed glioblastoma treated with enzyme inducing anti-epileptic drugs and standard radiation and temozolomide. J Neurooncol 103:325–332. https://doi.org/10.1007/s11060-010-0390-7
Bao J, Zhou N, Luo K et al (2014) In silico discovery of potential VEGFR-2 inhibitors from natural derivatives for anti-angiogenesis therapy. Int J Mol Sci 15:15994–16011. https://doi.org/10.3390/ijms150915994
Lu L, Saha D, Martuza RL et al (2015) Single agent efficacy of the VEGFR kinase inhibitor axitinib in preclinical models of glioblastoma. J Neurooncol 121:91–100. https://doi.org/10.1007/s11060-014-1612-1
Batchelor TT, Duda DG, Di Tomaso E et al (2010) Phase II study of cediranib, an oral pan-vascular endothelial growth factor receptor tyrosine kinase inhibitor, in patients with recurrent glioblastoma. J Clin Oncol 28:2817–2823. https://doi.org/10.1200/JCO.2009.26.3988
Lassman AB, Sepúlveda-Sánchez JM, Cloughesy T et al (2019) OS10.6 Infigratinib (BGJ398) in patients with recurrent gliomas with fibroblast growth factor receptor (FGFR) alterations: a multicenter phase II study. Neuro Oncol 30:21–22
Ma YS, Lin JJ, Lin CC et al (2018) Benzyl isothiocyanate inhibits human brain glioblastoma multiforme GBM 8401 cell xenograft tumor in nude mice in vivo. Environ Toxicol 33:1097–1104. https://doi.org/10.1002/tox.22581
Feng J, Yan PF, Zhao HY et al (2016) Inhibitor of nicotinamide phosphoribosyltransferase sensitizes glioblastoma cells to temozolomide via activating ROS/JNK signaling pathway. Biomed Res Int. https://doi.org/10.1155/2016/1450843
Galkin M, Jonas BA (2019) Enasidenib in the treatment of relapsed/refractory acute myeloid leukemia: an evidence-based review of its place in therapy. Core Evid 14:3–17. https://doi.org/10.2147/ce.s172912
Golub D, Iyengar N, Dogra S et al (2019) Mutant isocitrate dehydrogenase inhibitors as targeted cancer therapeutics. Front Oncol. https://doi.org/10.3389/fonc.2019.00417
Mellinghoff IK, Ellingson BM, Touat M et al (2020) Ivosidenib in isocitrate dehydrogenase 1-mutated advanced glioma. J Clin Oncol 38:3398–3406. https://doi.org/10.1200/JCO.19.03327
Usha T, Shanmugarajan D, Goyal AK et al (2018) Recent updates on computer-aided drug discovery: time for a paradigm shift. Curr Top Med Chem 17:3296–3307. https://doi.org/10.2174/1568026618666180101163651
Prieto-Martínez FD, López-López E, Eurídice Juárez-Mercado K, Medina-Franco JL (2019) Computational drug design methods—current and future perspectives. Silico Drug Des. https://doi.org/10.1016/b978-0-12-816125-8.00002-x
Mottini C, Napolitano F, Li Z et al (2021) Computer-aided drug repurposing for cancer therapy: approaches and opportunities to challenge anticancer targets. Semin Cancer Biol 68:59–74. https://doi.org/10.1016/j.semcancer.2019.09.023
Aljoundi AK, Agoni C, Fisaso AO, Soliman ME (2019) Turning to computer-aided drug design in the treatment of diffuse large B-cell lymphoma: has it been helpful? Anticancer Agents Med Chem 19:1325
Karplus M, McCammon JA (2002) Molecular dynamics simulations of biomolecules. Nat Struct Biol 9:646–652. https://doi.org/10.1038/nsb0902-646
Salmaso V, Moro S (2018) Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front Pharmacol 9:1–16. https://doi.org/10.3389/fphar.2018.00923
Hospital A, Goñi JR, Orozco M, Gelpí JL (2015) Molecular dynamics simulations: advances and applications. Adv Appl Bioinform Chem 8:37
Durrant J, McCammon JA (2011) Molecular dynamics simulations and drug discovery. BMC Biol 9:1–9
Dar AM, Mir S (2017) Molecular docking: approaches, types, applications and basic challenges. J Anal Bioanal Tech. https://doi.org/10.4172/2155-9872.1000356
Prieto-Martínez F, Medina-Franco J (2018) Molecular docking: current advances and challenges. TIP Rev Espec en Ciencias Quim Biol 21:65–87
Pan Y-L, Liu Y-L, Chen J-Z (2018) Computational simulation studies on the binding selectivity of 1-(1H-Benzimidazol-5-yl)-5-aminopyrazoles in complexes with FGFR1 and FGFR4. Molecules 23:767. https://doi.org/10.3390/molecules23040767
Lavecchia A, Di Giovanni C (2013) Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 20:2839–2860
Walters WP, Wang R (2020) New trends in virtual screening. J Chem Inf Model 60:4109–4111. https://doi.org/10.1021/acs.jcim.0c01009
Gimeno A, Ojeda-Montes MJ, Tomás-Hernández S et al (2019) The light and dark sides of virtual screening: what is there to know? Int J Mol Sci 20:1375. https://doi.org/10.3390/ijms20061375
Qing X, Lee XY, De Raeymaeker J et al (2014) Pharmacophore modeling: advances, limitations, and current utility in drug discovery. J Receptor Ligand Channel Res 7:81–92. https://doi.org/10.2147/JRLCR.S46843
Khedkar SA, Malde AK, Coutinho EC, Srivastava S (2007) Pharmacophore modeling in drug discovery and development: an overview. Med Chem 3:187–197
Kaserer T, Beck KR, Akram M et al (2015) Pharmacophore models and pharmacophore-based virtual screening: concepts and applications exemplified on hydroxysteroid dehydrogenases. Molecules 20:22799–22832. https://doi.org/10.3390/molecules201219880
Peter SC, Dhanjal JK, Malik V et al (2019) Quantitative structure-activity relationship (QSAR): modeling approaches to biological applications. In: Ranganathan S (ed) Encyclopedia of bioinformatics and computational biology. Elsevier, Amsterdam, pp 661–676
Wang T, Yuan XS, Wu MB et al (2017) The advancement of multidimensional QSAR for novel drug discovery—where are we headed? Expert Opin Drug Discov 12:769–784
Kar S, Leszczynski J (2020) Open access in silico tools to predict the ADMET profiling of drug candidates. Expert Opin Drug Discov 15:1473–1487. https://doi.org/10.1080/17460441.2020.1798926
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. https://doi.org/10.1093/bioinformatics/bti770
Tareq Hassan Khan M (2010) Predictions of the ADMET properties of candidate drug molecules utilizing different QSAR/QSPR modelling approaches. Curr Drug Metab 11:285–295. https://doi.org/10.2174/138920010791514306
Chandrasekaran B, Abed SN, Al-Attraqchi O et al (2018) Computer-aided prediction of pharmacokinetic (ADMET) properties. Elsevier Inc, Amsterdam
Lipinski CA (2000) Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44:235–249. https://doi.org/10.1016/S1056-8719(00)00107-6
Ya’u Ibrahim Z, Uzairu A, Shallangwa G, Abechi S (2020) Molecular docking studies, drug-likeness and in-silico ADMET prediction of some novel β-Amino alcohol grafted 1,4,5-trisubstituted 1,2,3-triazoles derivatives as elevators of p53 protein levels. Sci Afr 10:e00570. https://doi.org/10.1016/j.sciaf.2020.e00570
Stamos J, Sliwkowski MX, Eigenbrot C (2002) Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem 277:46265–46272. https://doi.org/10.1074/jbc.M207135200
Brozzo MS, Bjelić S, Kisko K et al (2012) Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization. Blood 119:1781–1788. https://doi.org/10.1182/blood-2011-11-390922
Fujikawa A, Sugawara H, Tanaka T et al (2017) Targeting PTPRZ inhibits stem cell-like properties and tumorigenicity in glioblastoma cells. Sci Rep 7:1–17. https://doi.org/10.1038/s41598-017-05931-8
Solca F, Dahl G, Zoephel A et al (2012) Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J Pharmacol Exp Ther 343:342–350. https://doi.org/10.1124/jpet.112.197756
Yosaatmadja Y, Silva S, Dickson JM et al (2015) Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed. J Struct Biol 192:539–544. https://doi.org/10.1016/j.jsb.2015.10.018
McTigue M, Murray BW, Chen JH et al (2012) Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc Natl Acad Sci USA 109:18281–18289. https://doi.org/10.1073/pnas.1207759109
Khan JA, Tao X, Tong L (2006) Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol 13:582–588. https://doi.org/10.1038/nsmb1105
Yen K, Travins J, Wang F et al (2017) AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov 7:478–493. https://doi.org/10.1158/2159-8290.CD-16-1034
Tripathi D, Imran S (2020) In-silico molecular docking study of novel derivatives of erlotinib in glioma. Asia-Pac J Mol Biol Biotechnol 28:34–38. https://doi.org/10.35118/apjmbb.2020.028.1.04
Yang RYC, Yang KS, Pike LJ, Marshall GR (2010) Targeting the dimerization of epidermal growth factor receptors with small-molecule inhibitors. Chem Biol Drug Des 76:1–9. https://doi.org/10.1111/j.1747-0285.2010.00986.x
Zhao M, Wang L, Zheng L et al (2017) 2D-QSAR and 3D-QSAR analyses for EGFR inhibitors. Biomed Res Int. https://doi.org/10.1155/2017/4649191
Panicker PS, Melge AR, Biswas L et al (2017) Epidermal growth factor receptor (EGFR) structure-based bioactive pharmacophore models for identifying next-generation inhibitors against clinically relevant EGFR mutations. Chem Biol Drug Des 90:629–636. https://doi.org/10.1111/cbdd.12977
Verma G, Khan MF, Akhtar W et al (2019) Pharmacophore modeling, 3D-QSAR, docking and ADME prediction of quinazoline based EGFR inhibitors. Arab J Chem 12:4815–4839. https://doi.org/10.1016/j.arabjc.2016.09.019
Hajalsiddig TTH, Osman ABM, Saeed AEM (2020) 2D-QSAR modeling and molecular docking studies on 1 H-Pyrazole-1-carbothioamide derivatives as EGFR kinase inhibitors. ACS Omega 5:18662–18674. https://doi.org/10.1021/acsomega.0c01323
Yadav M, Khandelwal R, Mudgal U et al (2019) Identification of potent VEGF inhibitors for the clinical treatment of glioblastoma, a virtual screening approach. Asian Pac J Cancer Prev 20:2681–2692. https://doi.org/10.31557/APJCP.2019.20.9.2681
Lutfiya AS, Priya S, Manzoor MAP, Hemalatha S (2019) Molecular docking and interactions between vascular endothelial growth factor (VEGF) receptors and phytochemicals: an in-silico study. Biocatal Agric Biotechnol 22:101424. https://doi.org/10.1016/j.bcab.2019.101424
Rathi E, Kumar A, Kini SG (2019) Molecular dynamics guided insight, binding free energy calculations and pharmacophore-based virtual screening for the identification of potential VEGFR2 inhibitors. J Recept Signal Transduct 39:415–433. https://doi.org/10.1080/10799893.2019.1690509
Agoni C, Ramharack P, Soliman MES (2018) Allosteric inhibition induces an open WPD-loop: a new avenue towards glioblastoma therapy. RSC Adv 8:40187–40197. https://doi.org/10.1039/C8RA08427K
Zhu Y, Liu A, Zhang X et al (2015) The effect of benzyl isothiocyanate and its computer-aided design derivants targeting alkylglycerone phosphate synthase on the inhibition of human glioma U87MG cell line. Tumor Biol 36:3499–3509. https://doi.org/10.1007/s13277-014-2986-6
Qian L, Zhu Y (2018) Computer-aided drug design and inhibitive effect of a novel nitrogenous heterocyclic compound and its mechanism on glioma U251 cells and breast cancer MCF-7 cells. Drug Des Dev Ther 12:1931–1939. https://doi.org/10.2147/DDDT.S168130
Yang B, Li X, He L, Zhu Y (2018) Computer-aided design of temozolomide derivatives based on alkylglycerone phosphate synthase structure with isothiocyanate and their pharmacokinetic/toxicity prediction and anti-tumor activity in vitro. Biomed Rep 8:235–240. https://doi.org/10.3892/br.2018.1051
Zhu YU, Han Y, Ma Y, Yang P (2018) Adme/toxicity prediction and antitumor activity of novel nitrogenous heterocyclic compounds designed by computer targeting of alkylglycerone phosphate synthase. Oncol Lett 16:1431–1438. https://doi.org/10.3892/ol.2018.8873
Zheng M, Sun W, Gao S et al (2017) Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1. Oncotarget 8:44255–44265. https://doi.org/10.18632/oncotarget.17464
Wang Y, Tang S, Lai H et al (2020) Discovery of novel IDH1 inhibitor through comparative structure- based virtual screening. Front Pharmacol 11:1–11. https://doi.org/10.3389/fphar.2020.579768
Zou F, Pusch S, Hua J et al (2018) Identification of novel allosteric inhibitors of mutant isocitrate dehydrogenase 1 by cross docking-based virtual screening. Bioorg Med Chem Lett 28:388–393. https://doi.org/10.1016/j.bmcl.2017.12.030
Zou F, Pusch S, Eisel J et al (2016) Identification of a novel selective inhibitor of mutant isocitrate dehydrogenase 1 at allosteric site by docking-based virtual screening. RSC Adv 6:96735–96742. https://doi.org/10.1039/c6ra21617j
Duan Z, Liu J, Niu L et al (2019) Discovery of DC_H31 as potential mutant IDH1 inhibitor through NADPH-based high throughput screening. Bioorg Med Chem 27:3229–3236. https://doi.org/10.1016/j.bmc.2019.05.040
Caravella JA, Lin J, Diebold RB et al (2020) Structure-based design and identification of FT-2102 (Olutasidenib), a potent mutant-selective IDH1 inhibitor. J Med Chem 63:1612–1623. https://doi.org/10.1021/acs.jmedchem.9b01423
Yang L, Pusch S, Jennings V et al (2019) Identification of new inhibitors of mutant isocitrate dehydrogenase 2 through molecular similarty-based virtual screening. Lett Drug Des Discov 16:861
Chandra N, Bhagavat R, Sharma E et al (2011) Virtual screening, identification and experimental testing of novel inhibitors of PBEF1/Visfatin/NMPRTase for glioma therapy. J Clin Bioinform 1:1–12. https://doi.org/10.1186/2043-9113-1-5
Ozgencil F, Eren G, Ozkan Y et al (2020) Identification of small-molecule urea derivatives as novel NAMPT inhibitors via pharmacophore-based virtual screening. Bioorg Med Chem 28:115217. https://doi.org/10.1016/j.bmc.2019.115217
Tanuma SI, Katsuragi K, Oyama T et al (2020) Structural basis of beneficial design for effective nicotinamide phosphoribosyltransferase inhibitors. Molecules 25:1–15. https://doi.org/10.3390/molecules25163633
Liu J, Wen Y, Gao L et al (2020) Design, synthesis and biological evaluation of novel 1H–1,2,4-triazole, benzothiazole and indazole-based derivatives as potent FGFR1 inhibitors viafragment-based virtual screening. J Enzyme Inhib Med Chem 35:72–84. https://doi.org/10.1080/14756366.2019.1673745
Kalman B, Szep E, Garzuly F, Post DE (2013) Epidermal growth factor receptor as a therapeutic target in glioblastoma. Neuro Mol Med 15:420–434. https://doi.org/10.1007/s12017-013-8229-y
Saleem H, Kulsoom Abdul U, Küçükosmanoglu A et al (2019) The TICking clock of EGFR therapy resistance in glioblastoma: target independence or target compensation. Drug Resist Updat 43:29–37. https://doi.org/10.1016/j.drup.2019.04.002
Loew S, Schmidt U, Unterberg A, Halatsch M-E (2012) The epidermal growth factor receptor as a therapeutic target in glioblastoma multiforme and other malignant neoplasms. Anticancer Agents Med Chem 9:703–715. https://doi.org/10.2174/187152009788680019
Hatanpaa KJ, Burma S, Zhao D, Habib AA (2010) Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance1. Neoplasia 12:675–684. https://doi.org/10.1593/neo.10688
Xu H, Zong H, Ma C et al (2017) Epidermal growth factor receptor in glioblastoma (Review). Oncol Lett 14:512–516. https://doi.org/10.3892/ol.2017.6221
Thorne AH, Zanca C, Furnari F (2016) Epidermal growth factor receptor targeting and challenges in glioblastoma. Neuro Oncol 18:914–918. https://doi.org/10.1093/neuonc/nov319
Roth P, Weller M (2014) Challenges to targeting epidermal growth factor receptor in glioblastoma: escape mechanisms and combinatorial treatment strategies. Neuro Oncol 16:viii14–viii19. https://doi.org/10.1093/neuonc/nou222
Kwatra MM (2019) A rational approach to target the epidermal growth factor receptor in glioblastoma. Curr Cancer Drug Targets 17:290–296. https://doi.org/10.2174/1568009616666161227091522
Choowongkomon K, Sawatdichaikul O, Songtawee N, Limtrakul J (2010) Receptor-based virtual screening of EGFR kinase inhibitors from the NCI diversity database. Molecules 15:4041–4054. https://doi.org/10.3390/molecules15064041
Silbey SS, American S, July N, Dobbin F (2016) Review: book reviews reviewed work (s): the common place of law stories from everyday life by Patricia, vol 105. University of Chicago Press, Chicago, pp 238–240. https://doi.org/10.1128/MCB.25.17.7734
Dawson JP, Berger MB, Lin C-C et al (2005) Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol 25:7734–7742. https://doi.org/10.1128/mcb.25.17.7734-7742.2005
Krcek R, Matschke V, Theis V et al (2017) Vascular endothelial growth factor, irradiation, and axitinib have diverse effects on motility and proliferation of glioblastoma multiforme cells. Front Oncol 7:1–11. https://doi.org/10.3389/fonc.2017.00182
Reardon DA, Turner S, Peters KB et al (2011) A review of VEGF/VEGFR-targeted therapeutics for recurrent glioblastoma. JNCCN J Natl Compr Cancer Netw 9:414–427. https://doi.org/10.6004/jnccn.2011.0038
Xu C, Wu X, Zhu J (2013) VEGF promotes proliferation of human glioblastoma multiforme stem-like cells through VEGF receptor 2. Sci World J. https://doi.org/10.1155/2013/417413
Chae YK, Ranganath K, Hammerman PS et al (2017) Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget 8:16052–16074. https://doi.org/10.18632/oncotarget.14109
Gallo LH, Nelson KN, Meyer AN, Donoghue DJ (2015) Functions of fibroblast growth factor receptors in cancer defined by novel translocations and mutations. Cytokine Growth Factor Rev 26:425–449. https://doi.org/10.1016/j.cytogfr.2015.03.003
Ardizzone A, Scuderi SA, Giuffrida D et al (2020) Role of fibroblast growth factors receptors (FGFRs) in brain tumors, focus on astrocytoma and glioblastoma. Cancers (Basel) 12:1–22. https://doi.org/10.3390/cancers12123825
Tonks NK (2006) Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol 7:833–846. https://doi.org/10.1038/nrm2039
Zhai YF, Beittenmiller H, Wang B et al (1993) Increased expression of specific protein tyrosine phosphatases in human breast epithelial cells neoplastically transformed by the Neu oncogene. Cancer Res 53:LP2272–LP2278
Müller S, Kunkel P, Lamszus K et al (2003) A role for receptor tyrosine phosphatase zeta in glioma cell migration. Oncogene 22:6661–6668. https://doi.org/10.1038/sj.onc.1206763
Ulbricht U, Brockmann MA, Aigner A et al (2003) Expression and function of the receptor protein tyrosine phosphatase zeta and its ligand pleiotrophin in human astrocytomas. J Neuropathol Exp Neurol 62:1265–1275. https://doi.org/10.1093/jnen/62.12.1265
Patel AP, Tirosh I, Trombetta JJ et al (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. https://doi.org/10.1126/science.1254257
**a Z, Ouyang D, Li Q et al (2019) The expression, functions, interactions and prognostic values of PTPRZ1: a review and bioinformatic analysis. J Cancer 10:1663–1674. https://doi.org/10.7150/jca.28231
Barr A (2010) Protein tyrosine phosphatases as drug targets: strategies and challenges of inhibitor development. Futur Med Chem 2:1563–1576
Benjamin DI, Cravatt BF, Nomura DK (2012) Global profiling strategies for map** dysregulated metabolic pathways in cancer. Cell Metab 16:565–577. https://doi.org/10.1016/j.cmet.2012.09.013
Piano V, Benjamin DI, Valente S et al (2015) Discovery of inhibitors for the ether lipid-generating enzyme AGPS as anti-cancer agents. ACS Chem Biol 10:2589–2597. https://doi.org/10.1021/acschembio.5b00466
Lee S, Urman A, Desai P (2019) Emerging drug profile: Krebs cycle and cancer: IDH mutations and therapeutic implications. Leuk Lymphoma 60:2635–2645. https://doi.org/10.1080/10428194.2019.1602260
Reitman ZJ, Yan H (2010) Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst 102:932–941. https://doi.org/10.1093/jnci/djq187
Huang J, Yu J, Tu L et al (2019) Isocitrate dehydrogenase mutations in glioma: from basic discovery to therapeutics development. Front Oncol 9:1–7. https://doi.org/10.3389/fonc.2019.00506
Huang LE (2019) Friend or foe-IDH1 mutations in glioma 10 years on. Carcinogenesis 40:1299–1307. https://doi.org/10.1093/carcin/bgz134
Ye D, Guan KL, **ong Y (2018) Metabolism, activity, and targeting of D- and L-2-hydroxyglutarates. Trends Cancer 4:151–165. https://doi.org/10.1016/j.trecan.2017.12.005
Waitkus MS, Diplas BH, Yan H (2016) Isocitrate dehydrogenase mutations in gliomas. Neuro Oncol 18:16–26. https://doi.org/10.1093/neuonc/nov136
Guo Q, Han N, Shi L et al (2019) NAMPT: a potential prognostic and therapeutic biomarker in patients with glioblastoma. Oncol Rep 42:963–972. https://doi.org/10.3892/or.2019.7227
Clark DE, Waszkowycz B, Wong M et al (2016) Application of virtual screening to the discovery of novel nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with potential for the treatment of cancer and axonopathies. Bioorg Med Chem Lett 26:2920–2926. https://doi.org/10.1016/j.bmcl.2016.04.039
Booth TC, Williams M, Luis A et al (2020) Machine learning and glioma imaging biomarkers. Clin Radiol 75:20–32. https://doi.org/10.1016/j.crad.2019.07.001
Valdebenito J, Medina F (2019) Machine learning approaches to study glioblastoma: a review of the last decade of applications. Cancer Rep 2:1–15. https://doi.org/10.1002/cnr2.1226
Macyszyn L, Akbari H, Pisapia JM et al (2016) Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol 18:417–425. https://doi.org/10.1093/neuonc/nov127
Niu B, Liang C, Lu Y et al (2020) Glioma stages prediction based on machine learning algorithm combined with protein-protein interaction networks. Genomics 112:837–847. https://doi.org/10.1016/j.ygeno.2019.05.024
Han L, Kamdar MR (2018) MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput. https://doi.org/10.1142/9789813235533_0031
Valdebenito S, D’Amico D, Eugenin E (2019) Novel approaches for glioblastoma treatment: focus on tumor heterogeneity, treatment resistance, and computational tools. Cancer Rep 2:1–13. https://doi.org/10.1002/cnr2.1220
Maddahi Y, Zareinia K, Gan LS et al (2016) Treatment of glioma using neuroArm surgical system. Biomed Res Int. https://doi.org/10.1155/2016/9734512
Hervey-Jumper SL, Li J, Lau D et al (2015) Awake craniotomy to maximize glioma resection: methods and technical nuances over a 27-year period. J Neurosurg 123:325–339. https://doi.org/10.3171/2014.10.JNS141520
Chiang GC, Kovanlikaya I, Choi C et al (2018) Magnetic resonance spectroscopy, positron emission tomography and radiogenomics-Relevance to glioma. Front Neurol 9:1–10. https://doi.org/10.3389/fneur.2018.00033
Kut C, Chaichana KL, ** J et al (2015) Detection of human brain cancer infiltration ex vivo and in vivo using quantitative optical coherence tomography. Sci Transl Med. https://doi.org/10.1126/scitranslmed.3010611
Pavlov V, Meyronet D, Meyer-Bisch V et al (2016) Intraoperative probe-based confocal laser endomicroscopy in surgery and stereotactic biopsy of low-grade and high-grade gliomas: a feasibility study in humans. Neurosurgery 79:604–611. https://doi.org/10.1227/NEU.0000000000001365
Liang D, Schulder M (2012) The role of intraoperative magnetic resonance imaging in glioma surgery. Surg Neurol Int. https://doi.org/10.4103/2152-7806.103029
Chang J, Narayana A (2010) Functional MRI for radiotherapy of gliomas. Technol Cancer Res Treat 9:347–358. https://doi.org/10.1177/153303461000900405
Salama GR, Heier LA, Patel P et al (2018) Diffusion weighted/tensor imaging, functional MRI and perfusion weighted imaging in glioblastoma-foundations and future. Front Neurol 8:1–11. https://doi.org/10.3389/fneur.2017.00660
Szymanski MD, Perry DW, Gage NM et al (2001) Magnetic source imaging of late evoked field responses to vowels: Toward an assessment of hemispheric dominance for language. J Neurosurg 94:445–453. https://doi.org/10.3171/jns.2001.94.3.0445
Frey D, Schilt S, Strack V et al (2014) Navigated transcranial magnetic stimulation improves the treatment outcome in patients with brain tumors in motor eloquent locations. Neuro Oncol 16:1365–1372. https://doi.org/10.1093/neuonc/nou110
Fang X, Liu M, Lu C et al (2019) Current status and potential application of navigated transcranial magnetic stimulation in neurosurgery: a literature review. Chinese Neurosurg J 5:1–7. https://doi.org/10.1186/s41016-019-0159-6
Alphandéry E (2018) Glioblastoma treatments: an account of recent industrial developments. Front Pharmacol 9:1–31. https://doi.org/10.3389/fphar.2018.00879
Nguyen NP, Nguyen ML, Vock J et al (2013) Potential applications of imaging and -guided radiotherapy for brain metastases and glioblastoma to improve patient quality of life. Front Oncol 3(NOV):1–7. https://doi.org/10.3389/fonc.2013.00284
Nandpuru HB, Salankar SS, Bora VR (2014) MRI brain cancer classification using support vector machine. In: 2014 IEEE Students' Conference on Electrical, Electronics and Computer Science. https://doi.org/10.1109/SCEECS.2014.6804439
Zakrzewski K, Jarzab M, Pfeifer A et al (2015) Transcriptional profiles of pilocytic astrocytoma are related to their three different locations, but not to radiological tumor features. BMC Cancer 15:1–16. https://doi.org/10.1186/s12885-015-1810-z
Prabhu VC (2021) http://www.aans.org/Patients/Neurosurgical-Conditions-and-Treatments/Astrocytomas-Tumors.Accessed 23 Aug 2021.
Khalid L, Carone M, Dumrongpisutikul N et al (2012) Imaging characteristics of oligodendrogliomas that predict grade. Am J Neuroradiol 33:852–857. https://doi.org/10.3174/ajnr.A2895
Romero FR, Zanini MA, Ducati LG et al (2012) Purely cortical anaplastic ependymoma. Case Rep Oncol Med 2012:1–4. https://doi.org/10.1155/2012/541431
Wilms G, Demaerel P, Sunaert S (2005) Intra-axial brain tumours. Eur Radiol 15:468–484. https://doi.org/10.1007/s00330-004-2555-2
Acknowledgements
We would like to acknowledge the School of Health Science, University of KwaZulu-Natal, Westville campus for financial assistance.
Funding
We would like to acknowledge the School of Health Science, University of KwaZulu-Natal, Westville campus for financial assistance.
Author information
Authors and Affiliations
Contributions
PP contributed towards literature surveys and preparation of manuscript. CA and MAAI contributed to proof-reading of the manuscript while MESS contributed as supervisor.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no financial and intellectual conflict of interest.
Ethical Approval
Not Applicable.
Consent to Participate
Not Applicable.
Consent for Publication
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Poonan, P., Agoni, C., Ibrahim, M.A.A. et al. Glioma-Targeted Therapeutics: Computer-Aided Drug Design Prospective. Protein J 40, 601–655 (2021). https://doi.org/10.1007/s10930-021-10021-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-021-10021-w