Abstract
In a survey of plant-parasitic nematodes in agricultural fields, cyst-forming nematodes were found in soil planted bamboo in Korea. The aim of this study was to identify the cyst nematodes based on morphological and molecular characteristics. As the results, the morphology and morphometrics of cysts and second-stage juveniles (J2s) were consistent with those of previous descriptions of Heterodera koreana. In phylogenetic analyses based on DNA sequences, these cyst nematodes were clustered together with clade of H. koreana in internal transcribed spacer (ITS) region, and large subunit D2-D3 segments (LSU D2-D3). These nematodes were clustered together with clade of H. koreana in cytochrome c oxidase subunit I (COI) gene, but a haplotype was different when compared with previous reported haplotypes (haplotype A-C) in Japan. This study showed these cyst nematodes were identified as H. koreana, and a new haplotype of H. koreana is distributed in Korea. We suggest that the new haplotype of H. koreana name as haplotype D.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant-parasitic nematodes (PPNs), which include over 4,100 species, interact with host crops (Decraemer & Hunt, 2006; Jones et al., 2013). Damage caused by PPNs has been estimated at 157 billion USD per year (Nicol et al., 2011). Among the PPNs, cyst nematodes are regarded as the most important PPNs worldwide, and contain eight genera as of 2021. Economic damage in many plants is caused by members of two genera, Heterodera and Globodera. The genus Heterodera contains some of the most important PPNs worldwide, and includes about 85 species including soybean cyst nematode (Heterodera glycines). The Heterodera species reported in Korea are as follows: soybean cyst nematode (H. glycines) on soybean in 1983 (Choi & Choi, 1983), Korean cyst nematode (H. koreana) on bamboo in 1992 (Vovlas et al., 1992), sugar beet cyst nematode (H. schachtii) on Kimchi-cabbage in 2011 (Kim et al., 2016), white soybean cyst nematode (H. sojae) on soybean in 2016 (Kang et al., 2016), clover cyst nematode (H. trifolii) on Kimchi-cabbage in 2017 (Mwamula et al., 2018), and rice cyst nematode (H. oryzae) on rice in 2020 (Mwesige et al., 2020).
H. koreana and H. sojae were first discovered in the Republic of Korea and were reported as novel cyst nematode species. Since then, the nematodes have been reported in other countries (Kang et al., 2016; Vovlas et al., 1992). H. koreana was described as Afenestrata koreana at first by Vovlas et al. (1992), but the genus Afenestrata was subsequently changed to Heterodera (Mundo-Ocampo et al., 2008). Because the genus Afenestrata was not classified as a specific clade and clustered together with Heterodera spp. in phylogenetic studies, synonymisation of Afenestrata with Heterodera was proposed (Mundo-Ocampo et al., 2008). Recently, bamboo has been cultivated as a crop for tea, and for seasoned vegetables in Korea, and damage by the Korean cyst nematode is expected. Since the Korean cyst nematode (H. koreana) was first reported, there have been no further findings or studies of the cyst nematode. A study of the cyst nematode based on its molecular characteristics is required because it is difficult to identify the Heterodera species using morphological and morphometrical characteristics.
In this study, we identified the cyst nematodes extracted from bamboo using morphological characteristics as well as molecular characteristics based on DNA barcoding genes such as LSU D2-D3 expansion segments, the ITS region, and the mitochondrial DNA COI gene. Furthermore, we identified the haplotype of the Korean cyst nematode based on the COI gene and named the new haplotype.
Materials and methods
Nematode isolation
Soil samples were collected from the rhizosphere of bamboo in three fields in Sacheon city, Gyeongsangnam-do in Korea, from soil at a depth of approximately 15 cm using a soil sampler (diameter 2 cm). The samples were labeled as BC348, DR437, and DR1256, respectively (Table 1). Cysts were extracted by the sieving method using 850 µm and 250 µm mesh sieves (Kang et al., 2016). After extraction, the cysts and J2s were transferred into a watch glass containing tap water using forceps and pipette under a stereomicroscope (MZ205; Leica, Wetzlar, Germany) and were kept at 4 °C until further use.
Morphological analysis
For light-microscopic observations, J2s were killed and fixed by addition of 80 °C FG 4:1 fixative (Southey, 1986). The nematodes were fixed for at least 24 h, then processed according to the Seinhorst method (Cid Del Prado Seinhorst, 1959; Vera & Subbotin, 2012). Specimens were mounted on Cobb slides and sealed with a paraffin ring and glycerin (Cobb, 1917). Vulval cones were cut under a stereomicroscope (M205; Leica, Wetzlar, Germany), and were transferred to glycerin on slide glasses. The nematodes were observed, measured, and photographed with the aid of a compound microscope (DM5000; Leica, Wetzlar, Germany) equipped with a microscope digital camera (DFC450; Leica, Wetzlar, Germany). The overall shape of the cysts, and the shape of the annule, vulval cone, head and lateral field were observed. The nematodes were identified morphologically based on the Heterodera species identification key authored by Subbotin et al. (2010).
Molecular analysis
To extract genomic DNA, each single cyst of three different populations was transferred to a slide-glass on a small drop of distilled water, opened and its contents crushed using a filter paper chip (2 mm × 2 mm) and forceps. Using forceps, the chip with the crushed eggs and J2s was transferred into a PCR tube containing 30 µl lysis buffer (sterilized triple distilled water, 1 M Tris–HCl, 10% Triton-X 100, 100 µg/ml Proteinase K, 2 M KCl, 1 M MgCl2) for extracting the nematode DNA (modified Iwahori et al., 2000). The tubes were incubated in a Thermal cycler (PTC-200, MJ Research, Alameda, CA, USA) at 60 °C for 1 h and 94 °C for 10 min.
Two ribosomal RNA fragments, i.e. the LSU D2-D3 segments, ITS regions, and COI gene of the mitochondrial genome were amplified. The primers for D2-D3 segment amplification were D2A (5’-ACAAGTACCGTGAGGGAAAGTTG-3’) and D3B (5’-TCGGAAGGAACCAGCTACTA-3’) (Subbotin et al., 2006). Primers for ITS amplification were TW81 (5’-GTTTCCGTAGGTGAACCTGC-3’) and AB28 (5’-ATATGCTTAAGTTCAGCGGGT-3’) (Subbotin et al., 2000). A primer set of JB3 (5’-TTTTTTGGGCATCCTGAGGTTTAT-3’) and JB5 (5’-AGCACCTAAACTTAAAACATAATGAAAATG-3’) for COI gene was used in the PCR reaction (Derycke et al., 2005).
The PCR conditions were as follows: pre-denaturation stage; 94 °C for 5 min, cycling stage (n = 40); denaturation at 94 °C for 1 min, annealing at 56 °C (D2-D3 segments), 58 °C (ITS region), and 57 °C (COI gene) for 1 min, respectively, and extension at 72 °C for 2 min. The final extension was carried out at 72 °C for 10 min. In order to verify the PCR amplicon, electrophoresis was performed using 0.5 × TAE buffer on 1% agarose gel. The amplicon was subsequently purified using a commercial PCR Purification Kit (Qiagen, Valencia, CA). All strands of the PCR amplicons were cycle-sequenced with an ABI PRISM BigDye Terminator version 1.1 Cycle Sequencing Kit and electrophoresed in each direction on an ABI Prism ABI 377 Genetic Analyzer (PE Applied Biosystems, USA). The newly obtained sequences were submitted to the GenBank database (Table 1).
Phylogenetic analysis
For phylogenetic study, the sequences of the three populations were compared with GenBank nematode sequences using the BLAST homology search program. The closest sequences were selected for phylogenetic analyses. Outgroup taxa for each dataset was chosen according to previous phylogenetic study for cyst-forming nematodes (Kang et al., 2016; Mwesige et al., 2020), Cryphodera brinkmani Karssen & van Aelst, 1999 and Meloidodera sikhotealiniensis Eroshenko, 1978 (De Luca et al., 2013; Subbotin et al., 2000, 2017). The newly obtained and published sequences for each gene were aligned using Clustal W with default parameters (Thompson et al., 1994). Sequence alignments were manually edited using BioEdit (Hall 1999). The alignment quality was examined by eye, and optimized manually by adjusting the ambiguous nucleotide positions. Models of base substitution were evaluated using MODELTEST3.7 combined with PAUP4.0 (Huelsenbeck & Ronquist, 2001; Posada & Crandall, 1998; Swofford, 2003). The Akaike-supported model, the base frequency, the proportion of invariable sites, and the gamma distribution shape parameters and substitution rates in the AIC were then used in phylogenetic analyses. Bayesian analysis was performed to confirm the tree topology for each gene separately using MrBayes 3.1.2 running the chain for 1 × 106 generations and setting the ‘burn-in’ at 2500 (Huelsenbeck & Ronquist, 2001). The MCMC (Markov Chain Monte Carlo) method was used within a Bayesian framework to estimate posterior probabilities of the phylogenetic trees (Larget & Simon, 1999), and generate a 50% majority- rule consensus tree. The posterior probabilities are given on appropriate clades. Trees were visualized using TreeView (Page, 1996).
Data of COI gene analysis
The sequences of the COI gene were assembled and aligned using MEGA version X (Kumer et al., 2018) with accession numbers LC202153-93, MW642452-4 and OL813218-9. DnaSP version 5.10.1 (Librado & Rozas, 2009) was used to carry out preliminary analyses of nucleotide polymorphism and haplotype variation, and to generate Arlequin haplotype files. We used PopART (Leigh & Bryant, 2015) to construct and examine median-joining haplotype networks (Bandelt et al., 1999), and Arlequin version 3.5.2.2 (Excoffier & Lischer, 2010) to estimate haplotype diversity and nucleotide diversity, and to test for genetic structure with an analysis of molecular variance (AMOVA).
Results
Morphological analysis
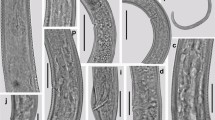
Morphological characters and morphometric features of cysts, and vulval cone of cysts and J2s were examined and measured for species identification. Lemon-shaped cysts, variable in size (377–903 µm) with distinct neck and vulval cones, were observed (Fig. 1A). The cuticle appeared light to dark brown in colour. The stylet and other pharyngeal structures were indistinct. A gelatinous egg sac was not observed and the cyst cuticle had an irregular zigzag pattern on mid-body. Vulval cone with lacking-fenestration was observed and covered with tuberculate pattern (Fig. 1B and C). J2s had a cylindrical body, tapering posteriorly, straight or slightly ventrally curved after fixation (Fig. 1D). The stylet had a length of 20–21 µm and was well developed, stylet-knobs were oviform (Fig. 1E and G), and the body length ranged from 454 to 545 µm (Table 2). The J2s had three incisures in the lateral field (Fig. 1F). Anus and hyaline part of tail were distinct, and the hyaline terminal section averaged 50.9 (45.5–55.0) µm long (Fig. 1H).
The morphological characterization of cyst, vulval cone and second-stage juvenile of Heterodera koreana. A: Lemon-shaped cyst; B-C: Vulval cone; D: Entire body of second-stage juveniles; E: Head region of second-stage juveniles; F: Lateral field; G: Anterior of second-stage juveniles; H: Tail region. (Scale bars: A = 100 µm, B-C = 50 µm, D = 100 µm, E = 20 µm, F = 10 µm and G-H = 50 µm)
Molecular and phylogenetic analysis
The LSU D2-D3 segments, ITS region, and COI gene of the mtDNA were amplified as indicated in the methodology section. The sequenced LSU D2-D3 segments, ITS region, and COI are 751–755, 902–908, and 424–425 bp, respectively. A BLASTn search on the LSU D2-D3 segments and ITS region revealed similarities with the Afenestrata group of Heterodera species such as H. Koreana and H. hainanensis. The highest match of the LSU D2-D3 segment sequences was H. koreana (LC202092), with 100% identities and no gaps. The ITS region results also revealed that the most similar species was H. koreana (KX640828), with 99.89% identities (901/902) and no insertions/deletions. In addition, a BLASTn search of H. koreana on the COI revealed high-scoring matches with H. koreana (LC202153), which is the species isolated from Phyllostachys nigra var. henonis in Iwate in Japan. The identities between the Korean population (this study) and H. koreana (LC202153) were 98.21% (385/392), with no insertions/deletions.
The molecular phylogenetic relationships of Korean populations of H. koreana were shown in Figs. 2, 3, and 4. The phylogenetic tree of the LSU D2-D3 segments of Heterodera species is shown in Fig. 2. The average nucleotide composition was as follows: 19.33% A, 21.47% C, 33.92 G and 25.28% T. Using Cryphodera brinkmani and Meloidodera sikhotealiniensis as the outgroup taxa, the molecular phylogeny strongly supported monophyly of Heterodera species. Phylogenetic tree inferred from the ITS region dataset is shown in Fig. 3. The average nucleotide composition was as follows: 19.17% A, 22.44% C, 29.04% G and 29.35% T. Three Korean populations of Heterodera species were close to H. koreana when C. brinkmani and M. sikhotealiniensis were used as the outgroup taxa. The results showed that the BC348, DR437, and DR1256 populations belong to the ‘Afenestrata’ group clade.
Phylogenetic relationships within population and species of Heterodera. Bayesian 50% majority rule consensus tree from two runs as inferred from the analysis of the D2-D3 of 28S rDNA gene sequences under the GTR + I + G model. Posterior probability values more than 50% are given in appropriate clades. Newly sequenced samples are indicated by bold font
Phylogenetic relationships within population and species of Heterodera. Bayesian 50% majority rule consensus tree from two runs as inferred from the analysis of the ITS rRNA gene sequences under the TVM + I + G model. Posterior probability values more than 50% are given in appropriate clades. Newly sequenced samples are indicated by bold font
Phylogenetic relationships between Heterodera species. Bayesian 50% majority rule consensus tree as inferred from the analysis of the COI gene sequence alignment under the TVM + I + G model. Posterior probabilities over 50% are given for appropriate clades. Newly obtained sequences are indicated by bold font
The phylogenetic tree based on the COI gene of mtDNA is shown in Fig. 4. The average nucleotide composition was as follows: 24.50% A, 8.68% C, 14.18 G and 52.64% T. Using Rotylenchus eximius and R. urmiaensis as the outgroup taxa, the molecular phylogenetic relationship of the dataset, which contains three Korean populations and previously registered data in NCBI, was closest to H. koreana. However, the three Korean populations were not clustered together with the Japanese haplotype of H. koreana, haplotype A-C, and were classified to a new clade of H. koreana.
Data of COI gene analysis
We identified four haplotypes (Haplotypes A–C, and Korean haplotype) in the 46 COI sequences (357 bp), and the dataset included 21 polymorphisms (Table 3). The network analysis showed a radial-shaped haplotype network with the most common Haplotypes A (48%) and B (43%) occupying a central position with the rest of the haplotypes differing by up to 13 substitutions (Fig. 5). Haplotypes A and C were found in Japan. Haplotype B was found in both Japan and the USA. The Korean haplotype was newly found in this study in Korea. We analyzed haplotype diversity and nucleotide diversity by regional populations. In the Japan populations, the haplotype diversity was 0.5317 ± 0.0319 and the nucleotide diversity was 0.0090 ± 0.0053. The Tajima’s D statistic was negative for the Japan populations (-0.68579). The Korean and USA populations had no diversity in the haplotype and the nucleotide, because there was no variation in the regional populations.
Pairwise FST, which is a fixation index between regional populations, was calculated to estimate the genetic differentiation that can be caused by the genetic structure. The Korean population had high fixation index values of 1.000 with the USA populations and 0.680 with the Japanese populations, indicating a differentiated genetic structure to the Korean population. On the other hand, the Japanese and USA populations had low differentiation, with a fixation index of 0.177. The AMOVA showed that variation by region was responsible for 60.46% of the total variation. The remaining 39.54% of the variance was explained by the variation among populations within region (7.76%) and variation within populations (31.78%) (Table 4).
Discussion
During a PPNs survey in 2020, three populations of Heterodera species were isolated from the rhizosphere of bamboo in the Republic of Korea. Three cyst-forming nematodes were identified as H. koreana using a morphological identification key for the Afenestrata sensu stricto group together with phylogenetic analysis (Table 2, Figs. 1 and 2). The genus Heterodera contains seven sensu stricto groups, which are the Afenestrata, Avenae, Cyperi, Goettingiana, Humuli, Sacchari and Schachtii groups (Subbotin et al., 2010). Bayesian trees inferred from LSU D2-D3 segments and the ITS region showed that H. koreana is related to the Afenestrata group (Figs. 2 and 3). The Afenestrata group includes the following seven species, which are H. africana, H. axonopi, H. bamboosi, H. hainanensis, H. koreana, H. orientalis and H. saccharophila (Mundo-Ocampo et al., 2008; Zhuo et al., 2013). Of these, three Heterodera species, which are H. bamboosi, H. koreana and H. hainanensis, have been recorded from bamboo in the world (Kaushal & Swarup, 1988; Vovlas et al., 1992; Zhuo et al., 2013), but the sequences of H. bamboosi were absent in GenBank. Nevertheless, H. koreana could be distinguish from H. bamboosi by the shorter body length in J2 (446 vs. 472 μm), and the vulval cone present in females (Subbotin et al., 2010).
Morphology and morphometrics of the cysts and the J2s of the three Korean populations were consistent with the described H. koreana in China (Wang et al., 2012). However, these populations differed from the original descriptions in the Republic of Korea and Japanese populations by the shorter cyst body length, the longer J2 body length, tail length, and the length of the hyaline region (Sekimoto et al., 2017; Vovlas et al., 1992). The ‘c’ value and ‘tail length’ in the Korean populations were inconsistent with the original description of the Iranian population (Maafi and Taheri, 2015). These differences in morphology could be explained as a result of intraspecific variation (Wang et al., 2012).
Recently, molecular and phylogenetic analysis based on DNA barcoding genes such as the LSU D2-D3 segments, ITS region, and the mtDNA COI gene have become very important to speed up and simplify the identification of animals including plant-parasitic nematodes. The COI gene is a very powerful DNA barcoding marker, and has been used in barcoding Heterodera species since 2005 (Blok & Powers, 2009; Derycke et al., 2005; Subbotin et al., 2015; Vovlas et al., 2015). However, our phylogenetic study based on the COI showed the haplotype of the Korean populations did not cluster together with Japanese populations (Fig. 4).
In a previous study, three COI haplotypes of H. koreana were found in Japan, and named as haplotypes A, B and C (Table 3). Haplotype A is dominant in Japan (Sekimoto et al., 2017). However, our study showed that a new haplotype of H. koreana was present in Korea, because the haplotype of the Korean populations was distinguished from the Japanese haplotype, and the AMOVA analysis also showed substantial regional differences (60.46%) between the Korean populations and Japanese populations, which was greater than that between the total populations (31.78%) (Table 4). Thus, we suggest that a new haplotype of H. koreana is present in the Republic of Korea and name it as haplotype D (Table 3 and Fig. 5).
To investigate associations between genetic haplotype and phenotype in the nematodes, we performed morphological comparisons between the Korean haplotype and the haplotypes reported in China and Japan. Our study showed that the whole length and hyaline portion in J2s in the Korean populations were similar to that of the Chinese population (Wang et al., 2012) (Table 2). However, our results showed that the Korean population had a longer J2 body length (499 ± 15.5 vs. 458 ± 17.3 μm) and a longer hyaline length of the J2 tail (51 ± 3.0 vs. 41 ± 3.2 μm) than those of the Japanese population (Sekimoto et al., 2017) (Table 2). Thus, we suggest that the differences in the haplotype of the COI gene sequence may affect the morphological differences within the species H. koreana. The studies on the association between the genotype of LSU D2-D3, ITS regions, COI, and morphological characteristics were conducted in marine nematodes. The results showed that highly divergent genotype clusters were accompanied by morphological differences (Derycke et al., 2008), and in the COI gene and ITS region particularly (Fonseca et al., 2008). Because the sequence of the Chinese haplotype is absent in NCBI Genbank, further study of the association between genetic haplotype and phenotype is required. In addition, the various haplotypes could be distributed in Republic of Korea due to the similar morphology between the original description of H. koreana in Republic of Korea and those of the Japanese population (Sekimoto et al., 2017; Vovlas et al., 1992). The haplotype may be associated with phenotypes of the nematodes like ecotype, and pathotype. Therefore, further study of correlations between the haplotypes of the nematodes (Haplotype A, B, C and, D) and phenotypes in bamboo, including a pathogenicity, is required.
References
Bandelt, H., Forster, P., & Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37–48.
Blok, V. C., & Powers, T. O. (2009). Biochemical and molecular identification. In Root-knot nematodes. (p. 98). CABI Publishing.
Choi, Y. E., & Choi, D. R. (1983). Survey on soybean parasitic nematodes. Korean Journal of Plant Protection, 22, 251–261.
Cobb, N. A. (1917). Notes on nemas. Contribution to a Science Nematology, 5, 117–128.
Decraemer, W., & Hunt, D. J. (2006). Structure and classification. In Plant nematology. (p. 3) CABI Publishing.
Derycke, S., Fonseca, G., Vierstraete, A., Vanfleteren, J., Vincx, M., & Moens, T. (2008). Disentangling taxonomy within the Rhabditis (Pellioditis) marina (Nematoda, Rhabditidae) species complex using molecular and morphological tools. Zoological Journal of the Linnean Society, 152, 1–15.
Derycke, S., Remerie, T., Vierstraete, A., Backeljau, T., Vanfleteren, J., Vincx, M., & Moens, T. (2005). Mitochondrial DNA variation and cryptic speciation within the free-living marine nematode Pellioditis marina. Marine Ecology Progress Series, 300, 91–103.
De Luca, F., Volvas, N., Lucarelli, G., Troccoli, A., Radicci, V., Fanelli, E., Catalapiedra-Navarrete, C., Palomares-Rius, J. E., & Castillo, P. (2013). Heterodera elachista the Japanese cyst nematode parasitizing corn in North Italy: Integrative diagnosis and bionomics. European Journal of Plant Pathology, 136, 857–872.
Excoffier, L., & Lischer, H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetic analyses under Linux and Windows. Molecular Ecology Resources, 10, 564–567.
Fonseca, G., Derycke, S., & Moens, T. (2008). Integrative taxonomy in two free-living nematode species complexes. Biological Journal of the Linnean Society, 94, 737–753.
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98NT. Nucleic Acids Symposium Serial, 41, 95–98.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Iwahori, H., Kanzaki, N., & Futai, K. (2000). A simple, polymerase chain reaction-restriction fragment length polymorphism aided diagnosis method for pine wilt disease. Forest Pathology, 30, 157–164.
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., Kikuchi, T., Manzanilla-lópez, R., Palomares-rius, J. E., Wesemael, W. M. L., & Perry, R. N. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology, 14, 946–961.
Kang, H., Eun, G., Ha, J., Kim, Y., Park, N., Kim, D., & Choi, I. (2016). New cyst nematode, Heterodera sojae n. sp. (Nematoda: Heteroderidae) from soybean in Korea. Journal of Nematology, 48, 280–289.
Karssen, G., & Aelst, A. V. (1999). Description of Cryphodera brinkmani n. sp. (Nematoda: Heteroderidae), a parasite of Pinus thunbergii Parlatore from Japan, including a key to the species of the genus Cryphodera Colbran, 1966. Nematology, 1(2), 121–130.
Kaushal, K. K., & Swarup, G. (1988). Two new cyst nematode species from India. Indian Journal of Nematology, 18, 299–306.
Kim, J. Y., Kim, T. H., Lee, Y. C., Chun, J. Y., Kern, E., Jung, J. W., & Park, J. K. (2016). Characterization of 15 microsatellite loci and genetic analysis of Heterodera schachtii (Nematoda: Heteroderidae) in South Korea. Biochemical Systematics and Ecology, 64, 97–104.
Kumer, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetic analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549.
Larget, B., & Simon, D. L. (1999). Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution, 16, 750–759.
Leigh, J. W., & Bryant, D. (2015). PopART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110–1116.
Librado, P., & Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452.
Maafi, Z. T., & Taheri, Z. M. (2015). First report of Korean cyst nematode, Heterodera koreana, parasitic on bamboo, Phyllostachys nigra, from Iran. Journal of Nematology, 47, 167–168.
Mundo-Ocampo, M., Troccoli, A., Subbotin, S. A., Cid, J. D., Baldwin, J. G., & Inserra, R. N. (2008). Synonymy of Afenestrata with Heterodera supported by phylogenetics with molecular and morphological characterisation of H. koreana comb. n. and H. orientalis comb. n. (Tylenchida: Heteroderidae). Nematology, 10, 611–632.
Mwamula, A. O., Ko, H., Kim, Y., Kim, Y., Lee, J., & Lee, D. W. (2018). Morphological and molecular characterization of Heterodera schachtii and the newly recorded cyst nematode, H. trifolii associated with Chinese cabbage in Korea. The Plant Pathology, 34, 297–307.
Mwesige, R., Kim, E., Park, E., & Ko, H. (2020). Morphological and molecular characterizations of Heterodera oryzae in Korea. Journal of Nematology, 52, 1–12.
Nicol, J. M., Turner, S. J., Coyne, D. L., den Nijs, L., Hockland, S., & Maafi, Z. T. (2011). Current nematode threats to world agriculture. Genomics and Molecular Genetics of Plant–Nematode Interactions, 21–43.
Page, R. D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Computer Applications in the Bioscience, 12, 357–358.
Posada, D., & Crandall, K. A. (1998). MODELTEST: Testing the model of DNA substitution. Bioinformatics, 14, 817–818.
Seinhorst, J. W. (1959). A rapid method for transfer of nematodes from fixative to anhydrous glycerin. Nematologica, 4, 67–69.
Sekimoto, S., Uehara, T., & Mizukubo, T. (2017). Morphological and molecular characterisation of Heterodera koreana (Vovlas, Lamberti & Choo, 1992) Mundo-Ocampo, Troccoli, Subbotin, Del Cid, Baldwin & Inserra, 2008 (Nematoda:Heteroderidae) from bamboo in Japan. Nematology, 19, 333–350.
Southey, J. F. (1986). Laboratory methods for work with plant and soil nematodes.
Subbotin, S. A., Mundo-Ocampo, M., & Baldwin, J. G. (2010). Systematics of cyst nematodes (Nematoda: Heteroderinae), Part B. Brill, Leiden, The Netherlands.
Subbotin, S. A., Sturhan, D., Chizhov, V. N., Vovlas, N., & Baldwin, J. G. (2006). Phylogenetic analysis of Tylenchida Thorne, 1949 as inferred from D2 and D3 expansion fragments of the 28S rRNA gene sequences. Nematology, 8, 455–474.
Subbotin, S. A., Waeyenberge, L., & Moens, M. (2000). Identification of cyst forming nematodes of the genus Heterodera (Nematoda: Heteroderidae) based on the ribosomal DNA-RFLPs. Nematology, 2, 153–164.
Subbotin, S. A., Vovlas, N., Yeates, G. W., Hallmann, J., Kiewnick, S., Chizhov, V. N., & Castillo, P. (2015). Morphological and molecular characterisation of Helicotylenchus pseudorobustus (Steiner, 1914) Golden, 1956 and related species (Tylenchida: Hoplolaimidae) with a phylogeny of the genus. Nematology, 17, 27–52.
Subbotin, S. A., Akanwari, J., Nguyen, C., del Prado, C., Vera, I., Chitambar, J., Inserra, R., & Chizhov, V. (2017). Molecular characterisation and phylogenetic relationships of cystoid nematodes of the family Heteroderidae (Nematoda: Tylenchida). Nematology, 19, 1065–1081.
Swofford, D. L. (2003). PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. Sinauer Associates, Sunderland.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Vera, I. C. D. P., & Subbotin, S. A. (2012). Belonolaimus maluceroisp. n. (Tylenchida: Belonolaimidae) from a tropical forest in Mexico and key to the species of Belonolaimus. Nematropica, 42, 201–210.
Vovlas, N., Lamberti, F., & Choo, H. Y. (1992). Description of Afenestrata koreana n. sp. (Nematoda: Heteroderinae), a parasite of bamboo in Korea. Journal of Nematology, 24, 553–559.
Vovlas, N., Vovlas, A., Leonetti, P., Liébanas, G., Castillo, P., Subbotin, S. A., & Palomares-Rius, J. E. (2015). Parasitism effects on white clover by root-knot and cyst nematodes and molecular separation of Heterodera daverti from H. trifolii. Eurpean Journal of Plant Pathology, 143, 833–845.
Wang, H. H., Zhuo, K., Zhang, H. L., & Liao, J. L. (2012). Heterodera koreana, a new record species from China. Acta Phytopathologica Sinica, 42, 551–555.
Zhuo, K., Wang, H., Ye, W., Peng, D., & Liao, J. (2013). Heterodera hainanensis n. sp. (Nematoda: Heteroderinae) from bamboo in Hainan Province, China – a new cyst nematode in the Afenestrata group. Nematology, 15, 303–314.
Acknowledgements
This research was supported by a fund (project no. FE0703-2023-01) from the National Institute of Forest Science and “Cooperative Research Program for Agriculture Science and Technology Department (Project No. PJ016606)” Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception, design, investigation, data analysis and interpretation, as well as drafting of the work. All authors have reviewed the manuscript and approved the final version of manuscript before submission.
Corresponding author
Ethics declarations
Ethics approval
All the authors certify that the work carried out in this research followed the principles of ethical and professional conduct have been followed.
Conflicts of interest
No potential conflict of interest relevant to this article was reported. Additionally, the authors certify that soil samplings did not involve any species endangered or protected in Republic of Korea.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kang, H., Ko, HR., Lim, YJ. et al. Haplotype diversity of Heterodera koreana (Tylenchida: Heteroderidae), affecting bamboo in Korea. Eur J Plant Pathol 169, 259–271 (2024). https://doi.org/10.1007/s10658-024-02823-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-024-02823-2