Abstract
It is often speculated that non-native invasive species undergo rapid changes in their phenotypic properties (i.e., traits) that provide adaptive advantage in their new environment. However, few studies have directly compared traits of invasive non-native species with their native counterparts to reveal whether such phenotypic changes occur, and which stages of initial introduction and subsequent invasion contribute to these shifts. We studied trait variation of an invasive tree, Pinus contorta, which is native to northwestern North America and invasive in the Patagonia region of South America (i.e., Argentina and Chile). Commercial plantations of P. contorta were introduced extensively in Patagonia from the 1970s onward, from an unknown seed origin within the Pacific Northwest, USA, where three sub-species are found, including subsp. contorta, latifolia, and murrayana. We employed a home-versus-away study approach, where we compared mean growth, defense, and reproduction trait values, and mean within-stand trait variation (Coefficient of Variation, CV) of Patagonia plantations, with the three native sub-species. We further compared mean traits, and trait CVs between invasive P. contorta and the Patagonia plantations from which they escaped. Patagonia plantations shared the most similar mean trait values with subsp. latifolia and murrayana, suggesting possible source populations. However, both mean trait values and trait CVs of Patagonia plantations differed from all three native sub-species, indicating potential founder effects, population bottlenecks, and/or plastic responses to their new environment that occurred during or after introduction. We also found evidence for selective change during invasion; however, these differences did not suggest growth traits were prioritized over defense traits, which was inconsistent with hypotheses that invaders exhibit an evolutionary trade-off between defense traits and growth traits. Our study highlights that processes occurring both at first introduction and establishment, as well as the subsequent invasion phase can influence the phenotype of successful invaders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the past two decades, plant functional trait analysis has developed into a major research theme in the fields of plant functional ecology and ecophysiology (Reich 2014). The trait-based approach has addressed many different topics, such as understanding the fundamental evolutionary trade-offs across plant species (Castorena et al. 2022), the functional linkage between dominant plant species and ecosystem properties and processes (Violle et al. 2007; Reich 2014; Diaz et al. 2016), or understanding what common characteristics are associated with successful plant invaders (van Kleunen et al. 2010). Regarding plant invaders, invasive plant species can sometimes have different traits than native species in the communities they invade (e.g. a higher average specific leaf area, SLA; Lake and Leishman 2004), or compared to their source population (e.g., plant height; Martín-Forés et al. 2023). These patterns suggest inherently higher growth rates are common characteristics of invaders (i.e., fast-resource acquisitive). A majority of trait studies, both within invasion ecology as well as other trait subjects, are focused at the across-species level (i.e. interspecific variation); however, there is growing recognition that intra-specific trait variation (ITV) can also be important, and contribute to community or ecosystem level functioning (Ackerly and Cornwell 2007; Fajardo and Piper 2011; Kumordzi et al. 2014; Siefert et al. 2015; Fajardo and Siefert 2018). It has been proposed that high phenotypic trait plasticity is a common characteristic of successful invaders, which may allow rapid adaptation to biotic or abiotic conditions in new environments (Geng et al. 2004; Davidson et al. 2011). Studies on ITV of invasive species are increasing rapidly, yet relatively few studies have analyzed ITV between a species’ native and non-native ranges, which has great potential to reveal underlying controls on ITV (Leger and Rice 2003; Martín-Forés et al. 2018a, 2018b).
Traits are properties of a phenotype, which are influenced by the environment (E), genetics (G), and the interplay between the two (i.e., G × E interactions). Hence, there are many mechanisms that can potentially influence invasive plant traits in new environments. Firstly, differences in environmental conditions between a species’ native and introduced ranges could cause trait differences (Geber and Griffen 2003; Barker et al. 2019). Secondly, considering that native populations can exhibit a wide range of genetic variability, introduction of genetically distinct individuals can cause introduced populations to exhibit different mean trait values, and lower trait variation, compared to their native counterparts, referred to as “founder effects” (Dlugosch and Parker 2008; Prentis et al. 2008). Invasive species often stem from small populations of initial colonizers, sometime chosen for their specific traits (e.g. fast growth or strait stems). Thus introduced populations generally are expected to have lower genetic diversity than populations within the native range. This may translate to lower phenotypic diversity; however, few studies have assessed this (Dlugosch and Parker 2008; Dlugosch et al. 2015).
In addition to the potential individual influences of genetics and environment on invasive species mean trait and trait variability values, interplays between environment and genetics (G × E) also offer potential explanations (Blossey and Notzold 1995; Richards et al. 2006; Prentis et al. 2008; Whitney and Gabler 2008). For example, plant populations may undergo rapid evolution of specific traits after arriving in a new environment (Prentis et al. 2008; Whitney and Gabler 2008), which may allow the population to be successful in the new environment. One such hypothesis, the Evolution of Increased Competitive Ability hypothesis (EICA), predicts that individuals with high growth or reproduction rates are favored over individuals who are well defended, due to the absence of enemies in the new environment (Blossey and Notzold 1995). Another way that G × E interactions can emerge in introduced populations is when different genotypes differ in their plastic response to their new environment. Some species or genotypes appear to be more able to respond quickly to a new environment by exhibiting greater plasticity (Richards et al. 2006). Consequently, more plastic individuals may have a higher chance of survival and successful reproduction, thus increasing the invasion potential of the species in a new environment (Martín-Forés et al. 2017).
There are clearly numerous hypotheses proposed to explain invasion success, and many of them may act simultaneously on different scales in time and space (Catford et al. 2009; Gurevitch et al. 2011). Many of these hypotheses suggest that key mean trait values and trait variability may change upon introduction to a new range. Despite the prevalence of hypotheses, relatively few studies have analyzed trait variation within and between species’ native and introduced environments (Leger and Rice 2003; Williams et al. 2008; Lamarque et al. 2015; Eyster and Wolkovich 2021). In this study, we evaluated mean trait values and mean trait variation values for 16 growth, defense, and reproduction traits from 32 sites (320 trees in total), spanning the native and an introduced range of the tree species Pinus contorta. Pinus contorta is native to Northwestern North America, where it consists of several subspecies, including contorta, latifolia and murrayana, which have been reported to differ in a variety of plant traits, such as needle and seed size, and cone shape (Lotan and Critchfield 1990). In the 1970s, P. contorta was widely introduced to the Patagonia region of South America. Because it reproduces at a very young age (ca. 5 years) and produces seeds that are easily dispersed by wind (Ledgard 2001; Gundale et al. 2014; Davis et al. 2019), subsequent generations have escaped plantations to become problematic invaders in the region. It is currently unknown which subspecies was introduced and invades the Patagonia region, and trait analysis may further help reveal which sub-species served as the origin for the Patagonian populations.
We tested the following hypotheses regarding ITV in P. contorta’s native and introduced ranges: (1) That mean trait values of the Patagonian plantations, which were the cohort originally introduced to the region, would differ from all subspecies in the native range, due to the potential influence of both “founder effects” and phenotypic plasticity responses to the Patagonia environment. (2) That trait variability (i.e., Coefficients of Variation, CV) is lower for introduced (i.e., plantations) and invasive populations compared to subspecies in the native range. We expected this because the founding population in Patagonia may have experienced a genetic bottleneck that could translate to low phenotypic diversity. (3) That the Patagonian plantation and invasive populations would exhibit trait differences, such as predicted by the EICA hypothesis. Specifically, we predicted invaders would exhibit an increase in growth-related traits (e.g., higher specific leaf area, or foliar nutrient contents), and a reduction in defense related traits (e.g., foliar lignin or phenolic contents). Testing these hypotheses in combination will yield new insights into how plant traits change upon introduction into a new environment, and a variety of processes that may underlie those patterns.
Materials and methods
Sites and field sampling
Our study consisted of 32 sites in total, including 24 in the Pacific Norwest, USA, and eight sites in the Patagonia region of Chile and Argentina (Fig. 1). Each site in North America consisted of 1 stand (24 in total), whereas in Patagonia, each site consisted of two paired stands, a plantation and an adjacent invasion (8 sites × two stand types = 16 stands). We sampled the North American sites in July 2018, and the South American sites in January 2019, such that sampling would cover a similar phenological period in each region. For North American sampling, the 24 sites we selected were divided among eight sites for each sub-species present in the Pacific Northwest, USA, including sub-species contorta, latifolia and murrayana (Fig. 1a). Each North American site consisted of a single mono-culture stand that was 40–60 years old. The remaining eight sites were located in the Patagonia region of Argentina and Chile (three and five sites, respectively; Fig. 1b). For each Patagonian site, adjacent P. contorta plantations and invasion stands were identified. The plantations were established in the 1970s (Fajardo and Gundale 2015), and the adjacent invading stands exhibited a clear invasion trajectory originating from the plantation (hereafter referred to as “Patagonian plantations” or “invasions front” stand types; Fig. 2). Once sites were selected, we randomly sampled traits from eight trees at each North American stand, as well as eight trees from each Patagonia plantation and invasion front (i.e., 8 trees within each stand × 5 stand types × 8 sites = 320 trees in total). Trees sampled at the “invasion front” in Patagonia consisted of sexually mature P. contorta trees furthest away from the plantations at each site (ca. 0.3–0.5 km; Fig. 2b).
The left panel a depicts sample sites of native Pinus contorta in Oregon and Washington State, USA. Pinus contorta subspecies are displayed using different symbols, including contorta (orange circles), murrayana (red squares), and latifolia (blue triangle). The right panel b depicts sample sites of introduced Pinus contorta in the Patagonia region of Chile and Argentina (green downward triangles)
For all trees sampled, we followed standard trait analysis protocols (Cornelissen et al. 2003), by collecting branch tips ca. 4 m high on the south or north side of the tree in the Northern or Southern Hemisphere, respectively (i.e., branches receiving full sun). Selected branches contained both brown and green cones, as well as new (i.e., current year) and fully expanded needles (i.e., previous year needles). Needles were collected from the previous year whorl, and green and brown cones were collected from the second- and third-year branch whorls (see further details below). The brown cones were fully expanded and open and had dispersed their seeds during the previous year. Green cones were less than two months from opening and dispersing their seeds. After collection, all samples were dried in an oven at 70 °C for 48 h. The heating process caused green cones to turn brown and fully open, releasing their seeds. After heating, leaf, cone, and seed traits were measured, as described further below.
Trait measurements
We measured a total of 16 traits from collected cones and needles (Tables 1, 2, 3, 4), which allowed us to analyze a trait “fingerprint” for each population. For each trait variable, multiple measurements were made for each selected tree in each stand, which allowed us to first compile average trait values per tree, and then subsequently compute averaged trait values for each stand, which served as the unit of replication (Cornelissen et al. 2003). Trait variables were based on needles, which relate to plant growth and defense, and seeds and cones, which are relevant for reproduction and dispersal. For needle traits, 20 needle fascicles (consisting of two needles) from each tree were weighed together and then divided by 20 to get average needle dry mass per fascicle per tree (Cornelissen et al. 2003). Then, 20 needle fascicles were scanned with a flatbed scanner (EPSON Perfection V800/V850 1.9 V3.93 3.9.3.2), and the resulting image was processed using WinRhizo 2016 to gain average needle length, and projected needle area. Surface areas of all the measured needles were then averaged for each tree with the unit mm2 per leaf. Leaf surface area and mass values were then used to compute specific leaf area (SLA = leaf area/leaf mass), which is considered a good indicator of plant growth and resource use strategy (Reich 2014), with high SLA values indicative of high growth rates. Further, high SLA values have been proposed to favor invasion success for non-native Pinus species (Grotkopp et al. 2002).
In addition to leaf physical measurements, we also measured a variety of leaf chemical traits, including polyphenolics, lignin, cellulose, nitrogen (N), carbon (C), and phosphorous (P) content. Here, we consider polyphenolics and lignin to represent defense traits, and foliar nutrient contents to serve as an indicator of growth capacity (Reich 2014). Thus, we interpret high values of polyphenols and lignin to equate with relatively higher defense, and high concentrations of N or P to be indicative of higher growth rates. For foliar cellulose and lignin analysis, we ground samples to 1 mm particle size using a ball mill. Lignin was measured with the acid-detergent fibre-sulphuric acid procedure (Helrich 1990), while cellulose was determined through nitric acid/acetic acid digestion, a subsequent reaction with anthrone, and measurement on a spectrophotometer at 620 nm, as described in Updegraff (1969). For C, N, and P analysis, samples were ground to a fine powder using a ball mill. Carbon and N content were measured by combustion and thermal conductivity detection using a Flash 1112 Elemental Analyzer (Thermo Scientific) linked to an Isotope Ratio Mass Spectrometer. Foliar P was measured by combustion and dissolution of ash in 1 M H2SO4, followed by phosphate detection, using automated molybdate colorimetry on a Lachat Quikchem 8500 analyzer. We further measured foliar phenolic concentrations using a modified version of the Prussian Blue Method (Stern et al. 1996; De Long et al. 2016; Grau-Andres et al. 2022). For this analysis, 50 mg of fine-ground foliar tissue was extracted in 20 ml of 50% methanol, and phenolic concentrations were compared against a catechin standard (Gundale et al. 2010; Piper et al. 2015).
Regarding cones, because oven drying caused green cones to turn brown and fully open, we combined data from brown and “green” cones for estimates of cone traits. Because green cones were collected at the end of their development period, they had reached their full size. We measured three cone traits, including length, width, and dry mass, from 20 cones per selected tree within each stand. While the relationship between these traits to function is not well established, we selected them because cone metrics have previously been shown to be diagnostic of the Pinus contorta subspecies (Critchfield 1957; Lotan and Critchfield 1990). Seeds were collected from the green cones after oven drying, and 20 seeds were randomly collected per tree for additional trait measurements. All selected seeds were weighed together, and then the weight was divided by the number of seeds, resulting in an average seed weight per tree. Using a scanner and WinRhizo software, we further measured the average seed length, and surface area.
To enable an evaluation of possible differences in traits between stand types, a mean stand value was calculated for each trait variable, by averaging each of the eight mean values derived from each sampled tree within each stand. We also calculated a coefficient of variation (CV) value for each trait in each stand, by first calculating the trait standard deviation (SD) among the eight measured trees within each stand, and then dividing the SD by the mean trait value from the same stand. This provided a measure of within-stand intraspecific trait variation (ITV) that could be compared across the population types, as opposed to across stand variation that would be sensitive to environmental variation.
Statistical analyses
We used RStudio (R Core Team 2019) for all univariate statistical analyses, and PRIMER package version 7 (Clarke and Gorley 2015) for multivariate data analyses. For all analyses, we considered the stand to be the unit of replication, which includes eight stands for each of the North American subspecies, and eight replicates for each Patagonia stand type (i.e., plantation and invasion fronts). We first tested whether there were differences in mean trait values and trait variation (CVs) between the Patagonia plantation stands and the three subspecies in the native range, using non-parametric multivariate analyses of variances (PERMANOVA) in PRIMER, where Bray–Curtis distances were applied to data matrices (Anderson 2001). To assure that the two site clusters in Patagonia (Argentina and Chile) were not significantly different from one-another, we compared their trait means and trait CVs using PERMANOVA (Table S1), which indicated they were not significantly different.
PERMANOVAs were also used to evaluate differences in mean trait values and trait variation of the introduced plantation trees and the invasive trees. Because the Patagonia stand types were paired within a site, we also included site as a random factor for this analysis. To test for homogeneity of multivariate dispersion, we used the permutation dispersion function (PERMDISP) in PRIMER, which we never found to be problematic. To graphically display PERMANOVA analysis of mean trait values and mean CV values, we performed two Canonical Analysis of Principal coordinates (CAP), one for mean trait values, and another for mean CV values. These CAP analyses included all five P. contorta stand categories (i.e., native subspecies contorta, latifolia, murrayana, and Patagonian plantation and invasion fronts), which allowed visualization of all categories relative to one another based on their mean trait or CV values.
We also followed all PERMANOVA tests with univariate analyses to evaluate whether any individual trait variables differed. This included one-way ANOVAs for each trait between subspecies contorta, latifolia, and murrayana, and Patagonia P. contorta plantations, and paired t-tests for each variable between the two Patagonia P. contorta stand types (i.e., plantations or invasions fronts). For all univariate analyses, we tested for assumption of normality using Shapiro–Wilk tests on the residuals of a linear model of the data, and Levene’s tests for the assumption of homogeneity of variance. If the data did not meet the assumptions, either a log- or BoxCox-transformation was done. When parametric assumptions were met, one-way ANOVA or t-tests were performed. When data could not be transformed to meet parametric assumptions, Welch’s one-way ANOVA tests or Kruskal–Wallis tests were used, when data were heterogeneous or non-normal, respectively (indicated in Tables 1, 2, 3, 4). If there was a difference between the groups, further post-hoc pairwise analyses were performed to examine which group differed. ANOVAs were followed by Tukey’s post-hoc tests, Welch’s One Way ANOVAs were followed by Games-Howell post-hoc tests, and Kruskal Wallis tests were followed using Dunn’s post-hoc tests. For all statistical analyses, a p-value ≤ 0.05 was considered as evidence of significant differences.
Results
Mean trait values
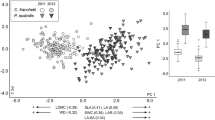
PERMANOVA indicated a significant difference (p ≤ 0.05) in multi-variate mean trait values among the four P. contorta stand categories (native subspecies contorta, latifolia, and murrayana, and Patagonia P. contorta plantations; Table 1). Post hoc pairwise PERMANOVA tests showed that multi-variate trait values of Patagonia plantations of Pinus contorta were the least similar to the North American Pinus contorta subspecies contorta, and relatively more similar to North American Pinus contorta subspecies latifolia and murrayana (Fig. 3a). Follow-up univariate post-hoc analyses showed that 14 of the 16 tested trait variables were significantly different among the four stand types (Table 2), except for needle P and N concentrations. In total, subspecies contorta was significantly different from subspecies latifolia and murrayana for 6 and 7 traits, respectively (Fig. 4). The native subspecies latifolia and murrayana were only significantly different from each other in needle cellulose content. Patagonian plantations were different from all three native subspecies for 4 trait variables (Fig. 4). These post-hoc analysis also indicated that Patagonian plantations had the greatest number of trait differences with native subspecies contorta, as well as latifolia, despite being relatively more similar in the multivariate analysis (11 traits different for each). In contrast, Patagonia plantations showed the fewest number of trait differences with sub-species murrayana (7 traits).
Results of Canonical Analysis of Principal coordinates on mean trait values a and within-stand trait co-efficient of variations b for sites of the five Pinus contorta stand categories. The analysis includes three native North American Pinus contorta sub-species, including contorta (yellow circles; C), latifolia (blue upward triangles; L), murrayana (red squares; M), and two introduced stand types, Patagonia Plantations (PP, green filled downward triangles) and Patagonia Invasion fronts (PI, green unfilled downward triangles). The δ2 values on the axes indicate the strength of the association between the data cloud of mean trait values and the hypothesis of differences between the five categories. In the lower left corner of each panel, within and between group similarities (%) are listed. An asterisk next to pairwise similarity values indicate when post hoc pairwise PERMANOVA analyses showed a significant difference (p < 0.05) in trait values between the two regions
Mean trait values for five Pinus contorta stand types, including native sub-species contorta (C, yellow), latifolia (L, blue), murrayana (M, Orange), and two introduced stand types, Patagonia Plantations (PP, left green) and Patagonia Invasion fronts (PI, right green). Black dots and error bars represent mean (± SE) trait values. Only trait variables exhibiting significant difference (p value < 0.05 from univariate analyses) are depicted. Different lower case letters (a–c) above means indicant significant differences between the first four categories, and uppercase letters (A, B) indicate significant differences between the two Patagonian stand types
Comparing the two stand types in Patagonia, we found no significant differences in multi-variate mean traits, which clustered tightly together in the CAP analysis (Table 2: Fig. 3a). Despite this, univariate comparisons showed three variables were different; foliar C and phenolic contents, and SLA were all higher in invading trees at the invasion front compared to trees in the plantations (Fig. 4).
Trait variation values
A significant difference in multi-variate trait CV values among the four categories was revealed by PERMANOVA (Table 3). Specifically, all four stand categories differed in their multi-variate trait CV values, except for native subspecies latifolia and murrayana (Fig. 3b). Of the 16 trait CV variables evaluated, univariate analysis showed a significant difference in eight traits among the categories (Table 4). Pairwise comparisons showed that native subspecies contorta differed significantly from latifolia and murrayana for four and three CV trait variables, respectively (Fig. 5). For those variables sub-species contorta generally displayed higher trait CV values. Subspecies murrayana differed from latifolia for only one trait variation value, seed surface area. Trait CVs of Patagonia plantations differed significantly from native sub-species contorta for three trait variables, from latifolia for one trait variable (cone width), and from murrayana for two trait variables (seed surface area and phenolic content). Regarding the two Patagonia stand types (plantations and invasion fronts), PERMANOVA and univariate comparisons never found significant differences in trait variation (Table 4).
Within stand trait coefficients of variation (CV) values for five Pinus contorta stand types, including native sub-species contorta (C, yellow), latifolia (L, blue), murrayana (M, Orange), and two introduced stand types, Patagonia Plantations (PP, left green) and Patagonia Invaders (PI, right green). Black dots and error bars represent mean CV (± SE) trait values. Only trait variables exhibiting significant differences (p-value < 0.05 from univariate analyses) are depicted. Different lower case letters (a or b) above means indicate significant differences between the first four categories, whereas no post hoc comparisons are depicted for the two Patagonian stand categories, because no PERMANOVA difference was detected
Discussion
We aimed to quantify variation between and within distinct populations of P. contorta, including three North American sub-species, and introduced plantations and invaders in the Patagonia region of South America, which have an unknown origin from the Pacific Northwest, USA. Our study showed substantial differences in mean trait values between the populations, as well as differences for within stand trait variability.
In support of our first hypothesis, multi-variate analysis showed that mean traits of Patagonia P. contorta plantations were dissimilar from all native sub-species. These differences included numerous traits for needles, cones, and seeds (Figs. 3, 4). While not all of these trait differences can be obviously associated with a fitness advantage, Patagonia plantations exhibited values of several traits considered to be more conservative, including higher concentrations of foliar phenolic defense compounds, and a lower SLA, suggesting a growth strategy that prioritizes defense over growth. Because Patagonia plantations were the first generation of trees introduced to the region, these differences cannot be explained by rapid evolution in response to their new environment. Alternatively, these traits differences could potentially be explained by plastic responses of the introduced trees to their new environment (Richards et al. 2006). For example, if the Patagonia environment was harsher or poorer in terms of soil resources than the East slope of the Cascades in Oregon and Washington, it is conceivable that genetically equivalent individuals would construct more conservative leaf architecture, as we observed. However, we have previously observed that the C:N ratios of soil are higher in the native range compared to Patagonia (Nuske et al. 2021), which is indicative of higher net nutrient turnover and availability in Patagonian soils. Thus, the lower C:N ratios we have observed in Patagonian soils does not align well with the more conservative needle traits observed in the region (Ordonez et al. 2009; Orwin et al. 2010; Laughlin et al. 2015). Some aspects of climate appear to be very similar between the Patagonia plantations and two of the native subspecies, including a very similar mean annual temperature with sampling sites of native subspecies latifolia and murrayana, and a very similar mean annual rainfall with subspecies murrayana (Table S2). This suggests that differences in climate are unlikely the explanation for the differences in trait values observed, especially regarding the differences between Patagonia plantations and native subspecies murrayana. However, an alternative to these “adaptive” explanations is that the observed differences are genetically controlled by founder effects, i.e., where genetically unusual individuals were purposely selected for introduction (Prentis et al. 2008), for example by selecting vigorous individuals in the native range that were more resistant to pathogens. This mechanism would imply that the observed trait differences are random or intentional, and may not have adaptive significance in their new environment.
While traits of Patagonia plantations differed from all native species, it is noteworthy that the degree of dissimilarity was not equal for all native sub-species. The Patagonia plantations had most similar trait values to both subspecies latifolia and murrayana, and most dissimilar with subspecies contorta. Further, univariate analysis showed murrayana exhibited the fewest trait differences with Patagonia plantations. The east slope of the Cascade Mountains in Oregon and Washington states, where subspecies latifolia and murrayana meet, share many similarities with the Patagonia region, including similar absolute latitudes, soils derived from volcanic ash, and climates influenced by mountain rain shadows, which are especially similar in terms of precipitation and temperature with the population area of subspecies murrayana (Table S2). While our study cannot distinguish between genetic and climatic controls on trait values, the higher similarity in trait values suggests the introduced Patagonia population may have originated from the east slope of the Cascades (i.e., where sub-species latifolia, murrayana, and their hybrids are found). This may have been an intentional decision by forest managers anticipating that these genotypes would yield viable phenotypes for the Patagonia region (Langdon et al. 2010; Gundale et al. 2014).
Regarding the North American subspecies, we also found numerous differences. Native subspecies contorta differed substantially from latifolia and murrayana, particularly regarding needle traits, where contorta exhibited lighter and shorter needles, smaller needle projected area, a lower C content, a higher lignin content, and a higher SLA. These differences are likely the result of genetic isolation and climate differences, given that subspecies contorta is separated from latifolia and murrayana by two mountain ranges (the Coastal and the Cascade Mountain ranges). Subspecies contorta is subjected to a coastal climate, which is characterized by less temperature variation, infrequent growing season frost, higher humidity, and higher and more frequent wind turbulence, whereas, subspecies latifolia and murrayana experience a more continental climate, including frequent growing season frost events. Our data are consistent with previous studies that have described sub-species contorta to have shorter and narrower needles compared to latifolia and murrayana (Critchfield 1957; Lotan and Critchfield 1990). Regarding cones, we found that mature cones of sub-species murrayana were significantly longer and wider than contorta, which corresponds with cone descriptions of Wheeler et al. (1983) and Koch (1996). However, previous descriptions of cone and seed traits of the different subspecies are sometimes contradicting (Critchfield 1957; Wheeler et al. 1983; Koch 1996), which may imply substantial spatial or inter-annual variation in these traits. These inconsistent patterns for cone traits suggest that leaf traits may serve as a more sensitive tool to distinguish P. contorta sub-species and populations. Regarding subspecies latifolia and murrayana, we found differences for only a single trait, needle cellulose content. This does not support previous descriptions that subspecies latifolia generally has longer and wider needles than murrayana (Lotan and Critchfield 1990). According to Wheeler et al. (1983), there are also differences in seed size between sub-species latifolia and murrayana; however, we found no support for this in our data. A possible explanation for this is that these two sub-species may overlap near the border of Oregon and Washington State (Fig. 1), which could cause substantial hybridization in our study area (Lotan and Critchfield 1990). The utility of traits to distinguish subspecies may therefore depend on the degree of population isolation and barriers to gene flow.
For our second hypothesis, we expected the introduced P. contorta plantations to exhibit lower trait variability compared to native subspecies due to a population bottleneck, which we evaluated by examining within stand trait coefficients of variation (CVs). Providing partial support for our hypothesis, multi-variate trait variation analysis showed that Patagonia plantations differed from North American P. contorta subspecies. Pairwise analyses showed that Patagonia plantations exhibited lower trait variation than at least one native subspecies for four traits (needle cellulose, needle phenolics, cone width, and cone weight), but higher trait variation for only one trait (seed surface area; Fig. 5), which provided further partial support for our hypothesis. It is possible that the traits with lower variation are under stronger genetic control, however, at this point there is little knowledge about which types of traits are more plastic versus constrained by genetics. Further, at this point there remains inconsistent evidence in the literature that exotic plant species exhibit lower trait variation. For example, Warwick et al. (1987) showed that trait variation of an introduced grass, Apera spica-venti, was not consistently lower than native grasses. Additionally, Dlugosch & Parker (2008) reviewed previously published literature and concluded that invasions do not show consistently lower variation in traits controlled by multiple genes in comparison to their native populations even though they often experience losses of genetic diversity, which is consistent with our findings. It is likely that substantial variation exists among exotic species in the degree to which genetic bottlenecks occur (Biedrzycka et al. 2014). Because P. contorta was intentionally introduced for commercial purposes, and the introductions occurred relatively recently (ca. 50 years ago), it is plausible that a small population of seeds served as the founding population. As such, this type of introduction history may be more likely to result in genetic bottlenecks compared to some other types of plant species, where repeated introduction events have occurred over longer time periods (Liao et al. 2020).
Regarding our third hypothesis, we expected to find differences in traits between plantations and invasion fronts of the Patagonian P. contorta population, which may serve as evidence for rapid evolution resulting in increased fitness in the non-native environment (Whitney and Gabler 2008). More specifically, the evolution of increased competitive ability hypothesis (EICA; Blossey and Notzold 1995) predicts that species may experience a rapid selection against defense traits (e.g., foliar phenolics; Lattanzio et al. 2006), and a rapid selection for traits that promote higher growth rates, such as a higher SLA and foliar N concentrations (Poorter 1989; Mitchell and Power 2003; Bossdorf et al. 2005). Our univariate analysis of individual traits showed that three of 16 mean trait values differed between Patagonia plantations and invaders, specifically C content, phenolic content, and SLA, all of which were observed to be higher among the invaders (Table 3, Fig. 4). While the higher SLA values we observed for invaders is consistent with the EICA hypothesis, the corresponding increase in phenolic content did not provide support for an evolutionary trade-off between growth and defense. Because the plantations we studied were established in the 1970s, and P. contorta trees can produce seeds after only 5 years (Langdon et al. 2010; Gundale et al. 2014), trees we sampled at the invasion front (i.e., the P. contorta trees located furthest away from the plantations) could represent anywhere between 1 and 10 generations that natural selection could have acted upon (Ledgard 2001). While it appears that some trait changes have already occurred among these generations, the shifts currently do not yet clearly indicate coordinated plant level shifts in allocation between growth and defense traits.
Conclusions
Pinus contorta is a widespread and problematic invader in Patagonia, as well as other regions such as New Zealand and Europe. Thus, better understanding where it comes from, and how its traits differ upon arrival to a new environment can help reveal why it may be successful as an invader. Our study provides several key insights into understanding its native origin, and adaptation to its new environment. First, our analysis of traits suggests that the P. contorta subsp. murrayana or murrayana-latifolia hybrids served as a source to the Patagonian population, due to their greatest trait similarity. At the same time, we also observed significant trait differences between Patagonian P. contorta and native latifolia and murrayana. This implies that P. contorta traits in Patagonia have been influenced by several non-mutually exclusive factors, including founder effects (i.e., genetically unusual individuals were introduced) or that trait plasticity has allowed its phenotype to adjust to the novel Patagonia environment. We also found some evidence of lower trait variation in the Patagonia population compared to the native sub-species, which suggests that a genetic bottleneck may have occurred during introduction, as a result of a small founding population. We further found a few trait differences between Patagonian plantations and invasions, which suggests that rapid evolution (i.e., change in allele frequencies) may be occurring in its new environment. However, these differences were not clearly consistent with the EICA hypothesis that predicts an evolutionary selection for growth traits at the expense of defense and suggests that trait shifts may be less coordinated than have been predicted. Taken together, our study highlights the substantial plant phenotypic variation that can occur across broad spatial and temporal scales, and across native and introduced ranges for a species. Further analysis is needed to clearly separate genetic versus environmental influences on this trait variation (e.g., genetic analysis or common garden experiments), but the current study provides a first step to indicate that processes occurring both at first introduction and at the subsequent invasion phase can influence intraspecific trait variability.
Data availability
Data presented in this manuscript are available upon reasonable request.
References
Ackerly DD, Cornwell WK (2007) A trait-based approach to community assembly: partitioning of species trait values into within- and among-community components. Ecol Lett 10:135–145
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46
Barker HL, Holeski LM, Lindroth RL (2019) Independent and interactive effects of plant genotype and environment on plant traits and insect herbivore performance: a meta-analysis with Salicaceae. Funct Ecol 33:422–435
Biedrzycka A, Zalewski A, Bartoszewicz M, Okarma H, Jedrzejewska E (2014) The genetic structure of raccoon introduced in Central Europe reflects multiple invasion pathways. Biol Invasions 16:1611–1625
Blossey B, Notzold R (1995) Evolution of increased competitive ability in invasive nonindigenous plants: a hypothesis. J Ecol 83:887–889
Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D (2005) Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144:1–11
Castorena M, Olson ME, Enquist BJ, Fajardo A (2022) Toward a general theory of plant carbon economics. Trends Ecol Evol 37:829–837
Catford JA, Jansson R, Nilsson C (2009) Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers Distrib 15:22–40
Clarke KR, Gorley RN (2015) PRIMER v7: User Manual/Tutorial (7). http://updates.primer-e.com/primer7/manuals/User_manual_v7a.pdf
Cornelissen JHC, Lavorel S, Garnier E, Diaz S, Buchmann N, Gurvich DE, Reich PB, ter Steege H, Morgan HD, van der Heijden MGA, Pausas JG, Poorter H (2003) A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust J Bot 51:335–380
Critchfield WB (1957) Geographic variation in Pinus contorta. Cambridge Press, Boston, Massachusetts
Davidson AM, Jennions M, Nicotra AB (2011) Do invasive species show higher phenotypic plasticity than native species and if so, is it adaptive? A meta-analysis. Ecol Lett 14:419–431
Davis KT, Callaway RM, Fajardo A, Pauchard A, Nunez MA, Brooker RW, Maxwell BD, Dimarco RD, Peltzer DA, Mason B, Ruotsalainen S, McIntosh ACS, Pakeman RJ, Smith AL, Gundale MJ (2019) Severity of impacts of an introduced species corresponds with regional eco-evolutionary experience. Ecography 42:12–22
De Long JR, Sundqvist MK, Gundale MJ, Giesler R, Wardle DA (2016) Effects of elevation and nitrogen and phosphorus fertilization on plant defence compounds in subarctic tundra heath vegetation. Funct Ecol 30:314–325
Diaz S, Kattge J, Cornelissen JHC, Wright IJ, Lavorel S, Dray S, Reu B, Kleyer M, Wirth C, Prentice IC, Garnier E, Bonisch G, Westoby M, Poorter H, Reich PB, Moles AT, Dickie J, Gillison AN, Zanne AE, Chave J, Wright SJ, Sheremet’ev SN, Jactel H, Baraloto C, Cerabolini B, Pierce S, Shipley B, Kirkup D, Casanoves F, Joswig JS, Gunther A, Falczuk V, Ruger N, Mahecha MD, Gorne LD (2016) The global spectrum of plant form and function. Nature 529:167-+
Dlugosch KM, Anderson SR, Braasch J, Cang FA, Gillette HD (2015) The devil is in the details: genetic variation in introduced populations and its contributions to invasion. Mol Ecol 24:2095–2111
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Eyster HN, Wolkovich EM (2021) Comparisons in the native and introduced ranges reveal little evidence of climatic adaptation in germination traits. Clim Change Ecol 2:100023
Fajardo A, Gundale MJ (2015) Combined effects of anthropogenic fires and land-use change on soil properties and processes in Patagonia, Chile. For Ecol Manage 357:60–67
Fajardo A, Piper FI (2011) Intraspecific trait variation and covariation in a widespread tree species (Nothofagus pumilio) in southern Chile. New Phytol 189:259–271
Fajardo A, Siefert A (2018) Intraspecific trait variation and the leaf economics spectrum across resource gradients and levels of organization. Ecology 99:1024–1030
Geber MA, Griffen LR (2003) Inheritance and natural selection on functional traits. Int J Plant Sci 164:S21–S42
Geng Y, Zhang W, Li B, Chen J (2004) Phenotypic plasticity and invasiveness of alien plants. Biodivers Sci 12:447–455
Grau-Andres R, Kardol P, Gundale M (2022) Trait coordination in boreal mosses reveals a bryophyte economics spectrum. Dryad
Grotkopp E, Rejmanek M, Rost TL (2002) Toward a causal explanation of plant invasiveness: seedling growth and life-history strategies of 29 pine (Pinus) species. Am Nat 159:396–419
Gundale MJ, Pauchard A, Langdon B, Peltzer DA, Maxwell BD, Nunez MA (2014) Can model species be used to advance the field of invasion ecology? Biol Invasions 16:591–607
Gundale MJ, Sverker J, Albrectsen BR, Nilsson MC, Wardle DA (2010) Variation in protein complexation capacity among and within six plant species across a boreal forest chronosequence. Plant Ecol 211:253–266
Gurevitch J, Fox GA, Wardle GM, Inderjit, and D. Taub. (2011) Emergent insights from the synthesis of conceptual frameworks for biological invasions. Ecol Lett 14:407–418
Helrich K (1990) Official methods of analysis of the association of official analytical chemists. Association of Official Analytical Chemists. Association of Official Analytical Chemists United States, Arlington, Virgina, pp 82–83
Koch P (1996) Lodgepole pine in North America. Forest Products Society, Madison, Wisconsin
Kumordzi BB, Nilsson MC, Gundale MJ, Wardle DA (2014) Changes in local-scale intraspecific trait variability of dominant species across contrasting island ecosystems. Ecosphere 5:1–7
Lamarque LJ, Lortie CJ, Porte AJ, Delzon S (2015) Genetic differentiation and phenotypic plasticity in life-history traits between native and introduced populations of invasive maple trees. Biol Invasions 17:1109–1122
Langdon B, Pauchard A, Aguayo M (2010) Pinus contorta invasion in the Chilean Patagonia: local patterns in a global context. Biol Invasions 12:3961–3971
Lattanzio V, Lattanzio VMT, Cardinali A (2006) Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects
Laughlin DC, Richardson SJ, Wright EF, Bellingham PJ (2015) Environmental filtering and positive plant litter feedback simultaneously explain correlations between leaf traits and soil fertility. Ecosystems 18:1269–1280
Ledgard N (2001) The spread of lodgepole pine (Pinus contorta, Dougl.) in New Zealand. For Ecol Manage 141:43–57
Leger EA, Rice KJ (2003) Invasive California poppies (Eschscholzia californica Cham.) grow larger than native individuals under reduced competition. Ecol Lett 6:257–264
Liao Z, Scheepens JF, Li Q, Wang W, Feng Y, Zheng Y (2020) Founder effects, post-introduction evolution and phenotypic plasticity contribute to invasion success of a genetically impoverished invader. Oecologia 192:2019
Lotan JE, Critchfield WB (1990) Pinus contorta Dougl. Ex Loud. Lodgepole Pine. In: Burns RMH (ed) Silvics of North America: Volume 1. Conifers. United States Department of Agriculture (USDA), Forest Service, Agriculture Handbook, pp 302–315
Martín-Forés I, Avilés M, Acosta-Gallo B, Breed MF, del Pozo A, de Miguel JM, Sánchez-Jardón L, Castro I, Ovalle C, Casado MA (2017) Ecotypic differentiation and phenotypic plasticity combine to enhance the invasiveness of the most widespread daisy in Chile, Leontodon saxatilis. Sci Rep 7:1546
Martín-Forés I, Acosta-Gallo B, Castro I, de Miguel JM, del Pozo A, Casado MA (2018a) The invasiveness of Hypochaeris glabra (Asteraceae): responses in morphological and reproductive traits for exotic populations. PLoS ONE 13:e0198849
Martín-Forés I, Casado MA, Castro I, del Pozo A, Molina-Montenegro MA, de Miguel JM, Acosta-Gallo B (2018b) Variation in phenology and overall performance traits can help to explain the plant invasion process amongst Mediterranean ecosystems. Neobiota:67–89
Martin-Fores I, Andrew SC, Guerin GR, Gallagher RV (2023) Linking the functional traits of Australian Acacia species to their geographic distribution and invasion status
Mitchell CE, Power AG (2003) Release of invasive plants from fungal and viral pathogens. Nature 421:625–627
Nuske SJ, Fajardo A, Nunez MA, Pauchard A, Wardle DA, Nilsson MC, Kardol P, Smith JE, Peltzer DA, Moyano J, Gundale MJ (2021) Soil biotic and abiotic effects on seedling growth exhibit context-dependent interactions: evidence from a multi-country experiment on Pinus contorta invasion. New Phytol 232:303–317
Ordonez JC, van Bodegom PM, Witte JPM, Wright IJ, Reich PB, Aerts R (2009) A global study of relationships between leaf traits, climate and soil measures of nutrient fertility. Glob Ecol Biogeogr 18:137–149
Orwin KH, Buckland SM, Johnson D, Turner BL, Smart S, Oakley S, Bardgett RD (2010) Linkages of plant traits to soil properties and the functioning of temperate grassland. J Ecol 98:1074–1083
Piper FI, Gundale MJ, Fajardo A (2015) Extreme defoliation reduces tree growth but not C and N storage in a winter-deciduous species. Ann Bot 115:1093–1103
Poorter H (1989) Causes and consequences of variation in growth rate and productivity of higher plants. In: Lambers H, Cambridge ML, Konings H, Pons TL (eds) Interspecific variation in relative growth rate: on ecological causes and physiological consequences. SPB Academic Publishing, The Hague, pp 45–68
Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ (2008) Adaptive evolution in invasive species. Trends Plant Sci 13:288–294
Reich PB (2014) The world-wide “fast-slow” plant economics spectrum: a traits manifesto. J Ecol 102:275–301
Richards CL, Bossdorf O, Muth NZ, Gurevitch J, Pigliucci M (2006) Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol Lett 9:981–993
Siefert A, Violle C, Chalmandrier L, Albert CH, Taudiere A, Fajardo A, Aarssen LW, Baraloto C, Carlucci MB, Cianciaruso MV, Dantas VD, de Bello F, Duarte LDS, Fonseca CR, Freschet GT, Gaucherand S, Gross N, Hikosaka K, Jackson B, Jung V, Kamiyama C, Katabuchi M, Kembel SW, Kichenin E, Kraft NJB, Lagerstrom A, Le Bagousse-**uet Y, Li YZ, Mason N, Messier J, Nakashizuka T, McC Overton J, Peltzer DA, Perez-Ramos IM, Pillar VD, Prentice HC, Richardson S, Sasaki T, Schamp BS, Schob C, Shipley B, Sundqvist M, Sykes MT, Vandewalle M, Wardle DA (2015) A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol Lett 18:1406–1419
Stern JL, Hagerman AE, Steinberg PD, Winter FC, Estes JA (1996) A new assay for quantifying brown algal phlorotannins and comparisons to previous methods. J Chem Ecol 22:1273–1293
Updegraff DM (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32:420–424
van Kleunen M, Weber E, Fischer M (2010) A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol Lett 13:235–245
Violle C, Navas ML, Vile D, Kazakou E, Fortunel C, Hummel I, Garnier E (2007) Let the concept of trait be functional! Oikos 116:882–892
Warwick SI, Thompson BK, Black LD (1987) Genetic variation in Canadian and European populations of the colonizing weed species Apera spica venti. New Phytol 106:301–317
Wheeler NC, Guries RP, Omalley DM (1983) Biosystematics of the genus Pinus, subsection contortae. Biochem Syst Ecol 11:333–340
Whitney KD, Gabler CA (2008) Rapid evolution in introduced species, “invasive traits” and recipient communities: challenges for predicting invasive potential. Divers Distrib 14:569–580
Williams JL, Auge H, Maron JL (2008) Different gardens, different results: native and introduced populations exhibit contrasting phenotypes across common gardens. Oecologia 157:239–248
Acknowledgements
This study was supported by a grant from the Swedish Research Council (Vetenskapsrådet; Project VR no. 2016-03819) awarded to MJG. AF was funded by Fondecyt 1190900. Special thanks also to Alice Trotel, Shoumo Khondoker, and Ilse van Duuren for laboratory assistance. The authors declare no conflicts of interest.
Funding
Open access funding provided by Swedish University of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gundale, M.J., Lindberg, L., Fajardo, A. et al. Functional traits differ across an invasive tree species’ native, introduced, and invasive populations. Biol Invasions 26, 2295–2311 (2024). https://doi.org/10.1007/s10530-024-03316-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-024-03316-3