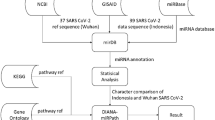
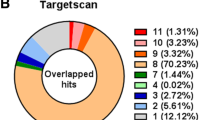
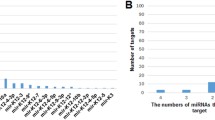
Abstract
Worldwide, many lives have been lost in the recent outbreak of coronavirus disease. The pathogen responsible for this disease takes advantage of the host machinery to replicate itself and, in turn, causes pathogenesis in humans. Human miRNAs are seen to have a major role in the pathogenesis and progression of viral diseases. Hence, an in-silico approach has been used in this study to uncover the role of miRNAs and their target genes in coronavirus disease pathogenesis. This study attempts to perform the miRNA seq data analysis to identify the potential differentially expressed miRNAs. Considering only the experimentally proven interaction databases TarBase, miRTarBase, and miRecords, the target genes of the miRNAs have been identified from the mirNET analytics platform. The identified hub genes were subjected to gene ontology and pathway enrichment analysis using EnrichR. It is found that a total of 9 miRNAs are deregulated, out of which 2 were upregulated (hsa-mir-3614-5p and hsa-mir-3614-3p) and 7 were downregulated (hsa-mir-17-5p, hsa-mir-106a-5p, hsa-mir-17-3p, hsa-mir-181d-5p, hsa-mir-93-3p, hsa-mir-28-5p, and hsa-mir-100-5p). These miRNAs help us to classify the diseased and healthy control patients accurately. Moreover, it is also found that crucial target genes (UBC and UBB) of 4 signature miRNAs interact with viral replicase polyprotein 1ab of SARS-Coronavirus. As a result, it is noted that the virus hijacks key immune pathways like various cancer and virus infection pathways and molecular functions such as ubiquitin ligase binding and transcription corepressor and coregulator binding.






Similar content being viewed by others
Data Availability
The data that support the findings of this study are available in NCBI Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra, reference number PRJNA736437, PRJNA737991. These data were derived from the following resources available in the public domain: PRJNA555016: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA736437/ and PRJNA394051: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA737991/
Change history
05 September 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10528-023-10511-9
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106. https://doi.org/10.1186/gb-2010-11-10-r106
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Avendaño-Ortiz J, Lozano-Rodríguez R, Martín-Quirós A, Maroun-Eid C, Terrón V, Valentín J, Montalbán-Hernández K, Ruiz de la Bastida F, García-Garrido MA, Cubillos-Zapata C et al (2020) Proteins from SARS-CoV-2 reduce T cell proliferation: a mirror image of sepsis. Heliyon 6(12):e05635. https://doi.org/10.1016/j.heliyon.2020.e05635
Aviv A (2020) Telomeres and COVID-19. FASEB J off Publ Fed Am Soc Exp Biol 34(6):7247–7252. https://doi.org/10.1096/fj.202001025
Bailey AL, Dmytrenko O, Greenberg L, Bredemeyer AL, Ma P, Liu J, Penna V, Winkler ES, Sviben S, Brooks E et al (2021) SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis. JACC Basic Transl Sci 6(4):331–345. https://doi.org/10.1016/j.jacbts.2021.01.002
Banaganapalli B, Al-Rayes N, Awan ZA, Alsulaimany FA, Alamri AS, Elango R, Malik MZ, Shaik NA (2021) Multilevel systems biology analysis of lung transcriptomics data identifies key miRNAs and potential miRNA target genes for SARS-CoV-2 infection. Comput Biol Med. 135:104570. https://doi.org/10.1016/j.compbiomed.2021.104570
di Bari I, Franzin R, Picerno A, Stasi A, Cimmarusti MT, Di Chiano M, Curci C, Pontrelli P, Chironna M, Castellano G et al (2021) Severe acute respiratory syndrome coronavirus 2 may exploit human transcription factors involved in retinoic acid and interferon-mediated response: a hypothesis supported by an in silico analysis. New Microbes New Infect 41:100853. https://doi.org/10.1016/j.nmni.2021.100853
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
Bermejo-Jambrina M, Eder J, Kaptein TM, van Hamme JL, Helgers LC, Vlaming KE, Brouwer PJM, van Nuenen AC, Spaargaren M, de Bree GJ et al (2021) Infection and transmission of SARS-CoV-2 depend on heparan sulfate proteoglycans. EMBO J 40(20):e106765. https://doi.org/10.15252/embj.2020106765
Buijsers B, Yanginlar C, de Nooijer A, Grondman I, Maciej-Hulme ML, Jonkman I, Janssen NAF, Rother N, de Graaf M, Pickkers P et al (2020) Increased plasma heparanase activity in COVID-19 Patients. Front Immunol 11:575047. https://doi.org/10.3389/fimmu.2020.575047
Cai C, Tang Y-D, Zheng C (2022) When RING finger family proteins meet SARS-CoV-2. J Med Virol 94(7):2977–2985. https://doi.org/10.1002/jmv.27701
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136(4):642–655. https://doi.org/10.1016/j.cell.2009.01.035
Catalanotto C, Cogoni C, Zardo G (2016) MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. https://doi.org/10.3390/ijms17101712
Cerezo-Magaña M, Bång-Rudenstam A, Belting M (2022) Proteoglycans: a common portal for SARS-CoV-2 and extracellular vesicle uptake. Am J Physiol Physiol 324(1):C76–C84. https://doi.org/10.1152/ajpcell.00453.2022
Chaouat AE, Achdout H, Kol I, Berhani O, Roi G, Vitner EB, Melamed S, Politi B, Zahavy E, Brizic I et al (2021) SARS-CoV-2 receptor binding domain fusion protein efficiently neutralizes virus infection. PLoS Pathog 17(12):e1010175. https://doi.org/10.1371/journal.ppat.1010175
Che Y, Jiang D, Zhang Y, Zhang Junqi XuT, Sun Y, Fan J, Wang J, Chang N, Wu Y et al (2022) Elevated ubiquitination contributes to protective immunity against severe SARS-CoV-2 infection. Clin Transl Med 12(12):e1103. https://doi.org/10.1002/ctm2.1103
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform 14(1):128. https://doi.org/10.1186/1471-2105-14-128
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G (2019) Trends in the development of miRNA bioinformatics tools. Brief Bioinform 20(5):1836–1852. https://doi.org/10.1093/bib/bby054
Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 92(4):418–423. https://doi.org/10.1002/jmv.25681
Chen J, Dai L, Kendrick S, Post SR, Qin Z (2022) The anti-COVID-19 drug remdesivir promotes oncogenic herpesvirus reactivation through regulation of intracellular signaling pathways. Antimicrob Agents Chemother. 66(3):e0239521. https://doi.org/10.1128/aac.02395-21
Chen J, Song J, Dai L, Post SR, Qin Z (2022b) SARS-CoV-2 infection and lytic reactivation of herpesviruses: a potential threat in the postpandemic era? J Med Virol 94(11):5103–5111. https://doi.org/10.1002/jmv.27994
Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8(Suppl 4):S11–S11. https://doi.org/10.1186/1752-0509-8-S4-S11
Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN et al (2020) SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183(4):1043-1057.e15. https://doi.org/10.1016/j.cell.2020.09.033
Croce L, Gangemi D, Ancona G, Liboà F, Bendotti G, Minelli L, Chiovato L (2021) The cytokine storm and thyroid hormone changes in COVID-19. J Endocrinol Invest 44(5):891–904. https://doi.org/10.1007/s40618-021-01506-7
Derosa L, Melenotte C, Griscelli F, Gachot B, Marabelle A, Kroemer G, Zitvogel L (2020) The immuno-oncological challenge of COVID-19. Nat Cancer 1(10):946–964. https://doi.org/10.1038/s43018-020-00122-3
Elliott J, Whitaker M, Bodinier B, Eales O, Riley S, Ward H, Cooke G, Darzi A, Chadeau-Hyam M, Elliott P (2021) Predictive symptoms for COVID-19 in the community: REACT-1 study of over 1 million people. PLoS Med 18(9):e1003777. https://doi.org/10.1371/journal.pmed.1003777
Farr RJ, Rootes CL, Rowntree LC, Nguyen THO, Hensen L, Kedzierski L, Cheng AC, Kedzierska K, Au GG, Marsh GA et al (2021) Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 17(7):e1009759. https://doi.org/10.1371/journal.ppat.1009759
Fong SJ, Dey N, Chaki J (2021). In: Dey N, Chaki J (eds) An introduction to COVID-19 BT - artificial intelligence for coronavirus outbreak. Springer, Singapore, pp 1–22
Girardi E, López P, Pfeffer S (2018) On the importance of host MicroRNAs during viral infection. Front Genet 9:439. https://doi.org/10.3389/fgene.2018.00439
Giuliani A, Matacchione G, Ramini D, Di Rosa M, Bonfigli AR, Sabbatinelli J, Monsurrò V, Recchioni R, Marcheselli F, Marchegiani F et al (2022) Circulating miR-320b and miR-483-5p levels are associated with COVID-19 in-hospital mortality. Mech Ageing Dev 202:111636. https://doi.org/10.1016/j.mad.2022.111636
Gordon S (2002) Pattern recognition receptors: doubling up for the innate immune response. Cell 111(7):927–930. https://doi.org/10.1016/S0092-8674(02)01201-1
Gottwein E, Cullen BR (2008) Viral and cellular MicroRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3(6):375–387. https://doi.org/10.1016/j.chom.2008.05.002
Grant AH, Estrada A III, Ayala-Marin YM, Alvidrez-Camacho AY, Rodriguez G, Robles-Escajeda E, Cadena-Medina DA, Rodriguez AC, Kirken RA (2021) The many faces of JAKs and STATs within the COVID-19 storm. Front Immunol 12:690477. https://doi.org/10.3389/fimmu.2021.690477
Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27(1):91–105. https://doi.org/10.1016/j.molcel.2007.06.017
Health WHO (2022) WHO Coronavirus (COVID-19) dashboard. [accessed 2022 Nov 30]. https://covid19.who.int/
Health WHO, Programme E, Panel EA, Preparedness IPC, Guidance IPC, Group D, Gdg IPC, Preparedness S, Plan R (2020) Transmission of SARS-CoV-2 : implications for infection prevention precautions. (July):1–10. [accessed 2022 Nov 12]. https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions
Huang H-Y, Lin Y-C-D, Li J, Huang K-Y, Shrestha S, Hong H-C, Tang Y, Chen Y-G, ** C-N, Yu Y et al (2020) miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res 48(D1):D148–D154. https://doi.org/10.1093/nar/gkz896
Inaba H, Aizawa T (2021) Coronavirus disease 2019 and the thyroid - progress and perspectives. Front Endocrinol (Lausanne). https://doi.org/10.3389/fendo.2021.708333
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23(1):3–20. https://doi.org/10.1038/s41580-021-00418-x
Jiang L, Tang B, Guo J, Li J (2022) Telomere length and COVID-19 outcomes: a two-sample bidirectional mendelian randomization study. Front Genet. https://doi.org/10.3389/fgene.2022.805903
Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309(5740):1577–1581. https://doi.org/10.1126/science.1113329
Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G et al (2018) DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res 46(D1):D239–D245. https://doi.org/10.1093/nar/gkx1141
Keikha R, Hashemi-Shahri SM, Jebali A (2021) The relative expression of miR-31, miR-29, miR-126, and miR-17 and their mRNA targets in the serum of COVID-19 patients with different grades during hospitalization. Eur J Med Res 26(1):75. https://doi.org/10.1186/s40001-021-00544-4
Khan MA-A-K, Sany MRU, Islam MS, Islam ABMMK (2020) Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 worldwide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front Genet. https://doi.org/10.3389/fgene.2020.00765
Koupenova M, Corkrey HA, Vitseva O, Tanriverdi K, Somasundaran M, Liu P, Soofi S, Bhandari R, Godwin M, Parsi KM et al (2021) SARS-CoV-2 initiates programmed cell death in platelets. Circ Res 129(6):631–646. https://doi.org/10.1161/CIRCRESAHA.121.319117
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11(9):597–610. https://doi.org/10.1038/nrg2843
Laporte M, Naesens L (2017) Airway proteases: an emerging drug target for influenza and other respiratory virus infections. Curr Opin Virol 24:16–24. https://doi.org/10.1016/j.coviro.2017.03.018
Letafati A, Najafi S, Mottahedi M, Karimzadeh M, Shahini A, Garousi S, Abbasi-Kolli M, Sadri Nahand J, Tamehri Zadeh SS, Hamblin MR et al (2022) MicroRNA let-7 and viral infections: focus on mechanisms of action. Cell Mol Biol Lett 27(1):14. https://doi.org/10.1186/s11658-022-00317-9
Li S, Zhang Y, Guan Z, Li H, Ye M, Chen X, Shen J, Zhou Y, Shi Z-L, Zhou P et al (2020) SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct Target Ther 5(1):235. https://doi.org/10.1038/s41392-020-00334-0
Li X, Cheng Z, Wang F, Chang J, Zhao Q, Zhou H, Liu C, Ruan J, Duan G, Gao S (2021) A negative feedback model to explain regulation of SARS-CoV-2 replication and transcription. Front Genet. https://doi.org/10.3389/fgene.2021.641445
Li X, Zhang Z, Wang Z, Gutiérrez-Castrellón P, Shi H (2022) Cell deaths: involvement in the pathogenesis and intervention therapy of COVID-19. Signal Transduct Target Ther 7(1):186. https://doi.org/10.1038/s41392-022-01043-6
Lokau J, Kleinegger F, Garbers Y, Waetzig GH, Grötzinger J, Rose-John S, Haybaeck J, Garbers C (2020) Tocilizumab does not block interleukin-6 (IL-6) signaling in murine cells. PLoS One 15(5):e0232612. https://doi.org/10.1371/journal.pone.0232612
Mohammed MEA (2021) The percentages of SARS-CoV-2 protein similarity and identity with SARS-CoV and BatCoV RaTG13 proteins can be used as indicators of virus origin. J Proteins Proteom 12(2):81–91. https://doi.org/10.1007/s42485-021-00060-3
Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, Atif SM, Hariprasad G, Hasan GM, Hassan MI (2020) Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys acta Mol basis Dis 1866(10):165878. https://doi.org/10.1016/j.bbadis.2020.165878
O’Sullivan JM, Gonagle DM, Ward SE, Preston RJS, O’Donnell JS (2020) Endothelial cells orchestrate COVID-19 coagulopathy. Lancet Haematol 7(8):e553–e555. https://doi.org/10.1016/S2352-3026(20)30215-5
Park A, Iwasaki A (2020) Type I and Type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 27(6):870–878. https://doi.org/10.1016/j.chom.2020.05.008
De Pasquale V, Quiccione MS, Tafuri S, Avallone L, Pavone LM (2021) Heparan sulfate proteoglycans in viral infection and treatment: a special focus on SARS-CoV-2. Int J Mol Sci. https://doi.org/10.3390/ijms22126574
Patil AH, Halushka MK (2021) miRge3.0: a comprehensive microRNA and tRF sequencing analysis pipeline. NAR Genom Bioinforma. https://doi.org/10.1093/nargab/lqab068
Pierce JB, Simion V, Icli B, Pérez-Cremades D, Cheng HS, Feinberg MW (2020) Computational analysis of targeting SARS-CoV-2, viral entry proteins ACE2 and TMPRSS2, and interferon genes by host MicroRNAs. Genes (Basel). https://doi.org/10.3390/genes11111354
Pijnappel EN, Suurmeijer JA, Groot Koerkamp B, Siveke JT, Salvia R, Ghaneh P, Besselink MG, Wilmink JW, van Laarhoven HWM (2021) Mandatory reporting measurements in trials for potentially resectable pancreatic cancer. Textbook of pancreatic cancer: principles and practice of surgical oncology. Springer, Cham, pp 107–118
Prevention C for DC (2020) COVID-19 and your health. Centers for Disease Control and Prevention. [accessed 2022 Sep 12]. https://www.cdc.gov/coronavirus/2019-ncov/your-health/index.html
Prevention C for DC (2022) Symptoms of COVID-19. [accessed 2022 Nov 12]. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html
Quiros-Roldan E, Amadasi S, Zanella I, Degli Antoni M, Storti S, Tiecco G, Castelli F (2021) Monoclonal antibodies against SARS-CoV-2: current scenario and future perspectives. Pharmaceuticals (Basel). https://doi.org/10.3390/ph14121272
Raskin S (2021) Genetics of COVID-19. J Pediatr (Rio J) 97(4):378–386. https://doi.org/10.1016/j.jped.2020.09.002
Riolo G, Cantara S, Marzocchi C, Ricci C (2021) miRNA Targets: from prediction tools to experimental validation. Methods Protoc. https://doi.org/10.3390/mps4010001
Rutter H, Parker S, Stahl-Timmins W, Noakes C, Smyth A, Macbeth R, Fitzgerald S, Freeman ALJ (2021) Visualising SARS-CoV-2 transmission routes and mitigations. BMJ 375:e065312. https://doi.org/10.1136/bmj-2021-065312
Santoso CS, Li Z, Rottenberg JT, Liu X, Shen VX, Fuxman Bass JI (2021) Therapeutic targeting of transcription factors to control the cytokine release syndrome in COVID-19. Front Pharmacol 12:673485. https://doi.org/10.3389/fphar.2021.673485
Sepe S, Rossiello F, Cancila V, Iannelli F, Matti V, Cicio G, Cabrini M, Marinelli E, Alabi BR, di Lillo A et al (2022) DNA damage response at telomeres boosts the transcription of SARS-CoV-2 receptor ACE2 during aging. EMBO Rep 23(2):e53658. https://doi.org/10.15252/embr.202153658
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B (2009) Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci USA 106(14):5761–5766. https://doi.org/10.1073/pnas.0808743106
Srivastava S, Garg I, Singh Y, Meena R, Ghosh N, Kumari B, Kumar V, Eslavath MR, Singh S, Dogra V et al (2023) Evaluation of altered miRNA expression pattern to predict COVID-19 severity. Heliyon 9(2):e13388. https://doi.org/10.1016/j.heliyon.2023.e13388
Szabo R, Bugge TH (2008) Type II transmembrane serine proteases in development and disease. Int J Biochem Cell Biol 40(6–7):1297–1316. https://doi.org/10.1016/j.biocel.2007.11.013
Szabo R, Bugge TH (2011) Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu Rev Cell Dev Biol 27:213–235. https://doi.org/10.1146/annurev-cellbio-092910-154247
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Teterina NL, Liu G, Maximova OA, Pletnev AG (2014) Silencing of neurotropic flavivirus replication in the central nervous system by combining multiple microRNA target insertions in two distinct viral genome regions. Virology 456–457:247–258. https://doi.org/10.1016/j.virol.2014.04.001
Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, Mildner A, Weaver SC, Ryman KD, Klimstra WB (2014) RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 506(7487):245–248. https://doi.org/10.1038/nature12869
Viner RM, Ward JL, Hudson LD, Ashe M, Patel SV, Hargreaves D, Whittaker E (2021) Systematic review of reviews of symptoms and signs of COVID-19 in children and adolescents. Arch Dis Child 106(8):802–807. https://doi.org/10.1136/archdischild-2020-320972
Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS Coronavirus. J Virol 94(7):e00127-20. https://doi.org/10.1128/JVI.00127-20
**ao F, Zuo Z, Cai G, Kang S, Gao X, Li T (2009) miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37:D105-10. https://doi.org/10.1093/nar/gkn851
Xu G, Wu Y, **ao T, Qi F, Fan L, Zhang S, Zhou J, He Y, Gao X, Zeng H et al (2022) Multiomics approach reveals the ubiquitination-specific processes hijacked by SARS-CoV-2. Signal Transduct Target Ther 7(1):312. https://doi.org/10.1038/s41392-022-01156-y
Yekta S, Shih I-H, Bartel DP (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304(5670):594–596. https://doi.org/10.1126/science.1097434
Yuan X, Yao Z, Wu J, Zhou Y, Shan Y, Dong B, Zhao Z, Hua P, Chen J, Cong Y (2007) G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am J Respir Cell Mol Biol 37(1):9–19. https://doi.org/10.1165/rcmb.2005-0345RC
Zhang Y, Li Y (2013) Regulation of innate receptor pathways by microRNAs. Sci China Life Sci 56(1):13–18. https://doi.org/10.1007/s11427-012-4428-2
Zhang H, Zheng H, Zhu J, Dong Q, Wang J, Fan H, Chen Y, Zhang X, Han X, Li Q et al (2021) Ubiquitin-modified proteome of SARS-CoV-2-infected host cells reveals insights into virus-host interaction and pathogenesis. J Proteome Res 20(5):2224–2239. https://doi.org/10.1021/acs.jproteome.0c00758
Zong Z, Wei Y, Ren J, Zhang L, Zhou F (2021) The intersection of COVID-19 and cancer: signaling pathways and treatment implications. Mol Cancer 20(1):76. https://doi.org/10.1186/s12943-021-01363-1
Acknowledgments
The authors would like to acknowledge the Department of Bioinformatics for providing high-performance computational facilities. RD would like to thank NSFDC, Ministry of Social Justice and Empowerment, Govt. of India, and UGC for providing a Junior Research Fellowship.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
The research idea was conceived, and the work was designed by RD and AV. The experiments and analyses were performed by RD. VSP and DP supported the sequence data analysis and image preparation. The experiments were supervised and approved by AV. All of the authors have equal contribution in drafting the manuscript. Finally, the manuscript was approved by all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
This study did not require any ethics approval, since the analysis was performed on publicly available biological data.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The author name Amouda Venkatesan has been corrected under the affiliation part and the equation “|log2-foldchange|≥ ± 1” under the ‘Methods’ section has been changed to “log2-foldchange ≥ ± 1”.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Das, R., Sinnarasan, V.S.P., Paul, D. et al. A Machine Learning Approach to Identify Potential miRNA-Gene Regulatory Network Contributing to the Pathogenesis of SARS-CoV-2 Infection. Biochem Genet 62, 987–1006 (2024). https://doi.org/10.1007/s10528-023-10458-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10458-x