Abstract
Background
Peroxisome proliferator-activated receptor delta (PPARδ) promotes inflammation and carcinogenesis in many organs, but the underlying mechanisms remains elusive. In stomachs, PPARδ significantly increases chemokine Ccl20 expression in gastric epithelial cells while inducing gastric adenocarcinoma (GAC). CCR6 is the sole receptor of CCL20. Here, we examine the role of PPARδ–mediated Ccl20/Ccr6 signaling in GAC carcinogenesis and investigate the underlying mechanisms.
Methods
The effects of PPARδ inhibition by its specific antagonist GSK3787 on GAC were examined in the mice with villin-promoter–driven PPARδ overexpression (PpardTG). RNAscope Duplex Assays were used to measure Ccl20 and Ccr6 levels in stomachs and spleens. Subsets of stomach-infiltrating immune cells were measured via flow cytometry or immunostaining in PpardTG mice fed GSK3787 or control diet. A panel of 13 optimized proinflammatory chemokines in mouse sera were quantified by an enzyme-linked immunosorbent assay.
Results
GSK3787 significantly suppressed GAC carcinogenesis in PpardTG mice. PPARδ increased Ccl20 level to chemoattract Ccr6+ immunosuppressive cells, including tumor-associated macrophages, myeloid-derived suppressor cells and T regulatory cells, but decreased CD8+ T cells in gastric tissues. GSK3787 suppressed PPARδ–induced gastric immunosuppression by inhibiting Ccl20/Ccr6 axis. Furthermore, Ccl20 protein levels increased in sera of PpardTG mice starting at the age preceding gastric tumor development and further increased with GAC progression as the mice aged. GSK3787 decreased the PPARδ-upregulated Ccl20 levels in sera of the mice.
Conclusions
PPARδ dysregulation of Ccl20/Ccr6 axis promotes GAC carcinogenesis by remodeling gastric tumor microenvironment. CCL20 might be a potential biomarker for the early detection and progression of GAC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the fifth most common malignancy and the fourth most lethal cancer worldwide, with a 5-year survival rate of 5%-10% in advanced stages [1, 2]. Novel GC therapies and reliable biomarkers for the early detection and progression of GC are urgently needed to improve the outcomes of GC patients.
Intensive research work has been performed to identify therapeutic targets for GC interventions, but progress has been limited, partially because of a lack of suitable in vivo preclinical models. Peroxisome proliferator-activated receptor delta (PPARδ, encoded by PPARD) is a nuclear transcriptional factor that regulates physiologic and pathophysiologic processes, especially those involved in cell stemness, inflammation, and tumorigenesis [3,4,5,6,7,8,9,10]. PPARδ is upregulated in human GC tissues, and its expression is associated with human GC grades and stages, suggesting PPARδ is a potential therapeutic target for GC. Moreover, we recently reported that transgenic PPARδ overexpression in villin-expressing gastric progenitor cells (VGPCs) [11] in mice (termed PpardTG mice) activates and transforms quiescent VGPCs and induces progression of gastric tumorigenesis from metaplasia to dysplasia, and finally to invasive intestinal-type gastric adenocarcinoma (GAC) that is associated with severe gastric chronic inflammation, faithfully recapitulating the pathogenic features of human intestinal-type GAC, the most common type of GC [8]. Chronic inflammation modulates tumor microenvironment (TME) to promote gastric tumor immune evasion and subsequently gastric carcinogenesis [12]. PPARδ expression in VGPCs triggered chronic gastric inflammation [8]; whether and how PPARδ modulates gastric TME to promote gastric carcinogenesis remains unknown.
The immunosuppressive TME (iTME) plays a pivotal role in promoting carcinogenesis [13, 14]. The populations of myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs) expand during tumorigenesis to promote iTME development and tumorigenesis including GC [15,16,17,18]. Our previous RNA-seq findings showed that the top three differentially altered pathways regulated by PPARδ were all related to immune modulatory mechanisms, and CCL20 was the chemokine most upregulated by transgenic PPARδ expression in VGPCs of PpardTG mice [8]. The sole receptor of CCL20 is CCR6 [19]. Accumulating data show that the alteration of CCL20/CCR6 axis promotes cancer progression in various types of tumors including GC [20, 21]. However, whether and how PPARδ upregulation of CCL20 in gastric epithelial cells orchestrates an iTME and subsequentially promotes GAC tumorigenesis in PpardTG mice remains unknown. Hence, in this study, we aimed to examine whether transgenic PPARδ expression in VGPCs orchestrates the gastric iTME to promote GAC tumorigenesis via upregulating the Ccl20/Ccr6 axis; whether specially targeted inhibition of PPARδ suppresses GAC tumorigenesis by inhibiting Ccl20/Ccr6 axis to significantly reverse the iTME; and whether circulating CCL20 in sera can serve as a potential biomarker for the early detection and tumor progression of GAC.
Materials and methods
Animals
PpardTG mice were produced from two independent founders (PpardTG-1 and PpardTG-2) that were generated at The University of Texas MD Anderson Cancer Center Genetically Engineered Mouse Facility by pronuclear injection of mouse Ppard expression construct under the control of a villin promoter (p12.4Kvill–Ppard) into fertilized FVB oocytes [22]. The two PpardTG mouse lines were found to exhibit similar PPARδ expression levels and display similar phenotypes [8, 22]. Therefore, all subsequent experiments were performed using randomly selected PpardTG-1 and PpardTG-2 mice, designated as PpardTG hereafter. The mice were housed with a dark/light cycle of 12 h, ambient temperature of 22 °C, and humidity of 30%-70%.
Mouse treatment with PPARδ antagonist GSK3787 diet and evaluation of gastric tumorigenesis
PpardTG mice and their sex- and gender- matched WT littermates at age 6–8 weeks were fed either control diet (#TD.110161: 2019 Teklad Global 19% Protein Rodent Diet, Envigo) or the same control diet but with adding GSK3787 (#G7423, Sigma-Aldrich; customized diet, Envigo, 200 mg/kg) for 44 weeks (n = 6–10 mice per group). Then, the mice were euthanized, and the stomach of each mouse was removed, weighed, photographed, and grossly inspected for tumor formation. Half of the stomach from each mouse was harvested for RNA and protein analyses, and the other half was put in 10% neutral formalin for further sectioning analysis.
Mouse stomach tissue histology
Formalin-fixed gastric tissue samples were embedded in paraffin and sectioned onto slides at 5 μm thick and then stained with hematoxylin and eosin. Digital hematoxylin and eosin staining slides for mouse gastric tissues were scanned with Aperio AT2 (Leica Biosystems), and the scanned images were captured with Aperio ImageScope software (v12.3.3.5048). Histologic assessment for gastric tumor lesions was performed with the support of an experienced rodent pathologist in the MD Anderson veterinary pathology services facility.
In situ hybridization staining of RNAscope duplex assay
In situ hybridization staining of RNAscope Duplex Assay was performed according to the manufacturer’s manual (RNAscope 2.5 HD Duplex assay, #322500-QKG, Bio-Techne ACDBio). Briefly, the freshly cut 5 µm-sections were deparaffinized and treated with 3% H2O2. The slides were then boiled in RNAscope specific target retrieval reagent (Bio-Techne ACDBio), washed with 100% ethanol, dried, and incubated with protease reagent in the humidity control tray in the HybEZ oven (Bio-Techne ACDBio). Next, the hybridization was performed in the HybEZ oven using the following probes: mouse Ccl20 (#434051, Bio-Techne ACDBio) with mouse villin (#463301-C2, Bio-Techne ACDBio) or with mouse Ccr6 (#424461-C2, Bio-Techne ACDBio). After the hybridization, the slides were washed, and the signals were amplified by using RNAscope 2.5 HD Duplex Detection Kit (#322500, Bio-Techne ACDBio), followed by Mayer’s hematoxylin staining. RNA in situ hybridization intensity was scored as follows: the red dots as positive staining per cell for each slide were counted under microscope with 10X magnification and scored and averaged for five random fields as follows: 0 (no staining or < 1 dot/10 cells), 1 + (1–3 dots/cell), 2 + (4–10 dots/cell, very few dot clusters), 3 + (> 10 dots/cell, and < 10% dot clusters), or 4 + (> 10 dots/cell, and ≥ 10% dot clusters) according to the manufacturer’s guideline (SOP 45–006, Bio-Techne ACDBio).
Statistical analysis
Statistical significance was determined by the unpaired Student t-test or analysis of variance (one-way ANOVA with Tukey’s multiple comparisons test or two-way ANOVA with Šídák’s multiple comparisons test). Kaplan–Meier survival analysis and the log-rank test were used to compare survival outcomes. The statistical significance of the correlation of two factors was determined by Spearman correlation analysis. All tests were two-sided, and significance was defined at P < 0.05. Data were analyzed using SAS software, version 9.4 (SAS Institute) or GraphPad Prism 9 (GraphPad Software). Values presented are mean ± standard error of the mean (SEM) unless otherwise specified (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Results
PPARδ expression is negatively associated with overall survival (OS) and progression-free survival (PFS) in a large cohort of GC patients
PPARδ is upregulated in GC tissues and its expression is associated with GC grades and stages in patients with GC [8]. To further evaluate the impact of PPARδ on the patients’ survival, we searched a large cohort of GC patients in the Kaplan–Meier Plotter public database [23] and found that PPARδ mRNA expression levels measured with a specific PPARδ probe (208044_s_at) were negatively associated with the GC patients’ OS and PFS: among the 875 evaluated GC patients, those with low PPARδ mRNA expression had longer median OS (89.4 months [n = 318]) and PFS (32.4 months [n = 363]) than those with high PPARδ mRNA expression (OS: 23.5 months [n = 557] and PFS: 12.2 months [n = 277]) (Fig. 1a, b). Similarly, among the 320 patients with GAC (intestinal type), those with low PPARδ mRNA expression had longer median OS (123.8 months [n = 188]) and PFS (93.2 months [n = 141]) than those with high PPARδ mRNA expression (OS: 25.9 months [n = 132] and PFS: 19.4 months [n = 122]) (Fig. 1c, d).
Kaplan–Meier plots of OS and PFS for GAC (all types) and GAC (intestinal-type) patients with low vs. high PPARδ expression. a, b OS and PFS for patients with GAC of all types. c, d OS and PFS for patients with GAC (intestinal type). P values were calculated by log-rank test. HR (95% confidence interval) was from Cox regression using the high expression group as reference. HR hazard ratio
PPARδ-specific antagonist GSK3787 suppresses GAC progression in mice
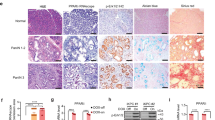
Our previous finding that PPARδ overexpression in VGPCs in mice induces large and invasive GAC accompanied with severe gastric chronic inflammation [8] prompted us to explore whether specifically targeting PPARδ is effective for GAC interventions, given that PPARδ antagonists have been developed. GSK3787 is a synthetic and irreversible antagonist of PPARδ that covalently binds to a cysteine residue in the ligand-binding domain of PPARδ [24]. In this study, we fed PpardTG mice at age 6–8 weeks the GSK3787 (200 mg/kg) or a control diet for 44 weeks. We found that this GSK3787 treatment significantly inhibited GAC carcinogenesis by decreasing stomach weights and inhibiting inflammatory cells’ infiltration, and GAC progression (Fig. 2a–c). A total of 7 of 10 PpardTG mice fed the control diet developed locally invasive GAC, but none of the sex- and age-matched PpardTG mice fed the GSK3787 diet had invasive GAC (Fig. 2c). The weights of spleens from PpardTG mice were significantly higher than those in control WT mice, but GSK3787 treatment had no effects on spleen weights in either WT or PpardTG mice (Fig. 2d). To assess the side effects of this long-term treatment with GSK3787, we evaluated liver and kidney function in the mice used for Fig. 2 by measuring serum levels of total protein, albumin, globulin, ALP, ALT and AST to monitor liver function and BUN and creatinine to monitor kidney function. While, unsurprisingly, PpardTG mice had lower serum protein levels, higher ALP and higher ALT levels than WT mice (Table S1), GSK3787 had no significant effects on liver or kidney function for WT and PpardTG mice (WT–Ctrl vs WT–GSK3787 or PpardTG–Ctrl vs PpardTG–GSK3787), indicating that GSK3787 was well tolerated in this preclinical study. These results further demonstrate the critical role of PPARδ in GAC tumorigenesis and suggest potential translational implications of targeting altered PPARδ for GAC interventions.
PPARδ–specific antagonist GSK3787 suppressed GAC progression in PpardTG mice. PpardTG mice and WT littermates at age 6 weeks (6–10 mice per group) were fed a GSK3787 (200 mg/kg) or control (Ctrl) diet for 44 weeks and then euthanized. a Gross examination of the stomachs. b Weights of stomachs. c Photomicrographs of hematoxylin and eosin–stained sections of the stomachs showing locally invasive adenocarcinoma into the muscle layers of stomach walls and marked transmural infiltrations of many lymphocytes in PpardTG mice fed a control diet in comparison with non-invasive tumors in PpardTG mice fed a GSK3787 diet. None of the WT mice developed gastric tumors. Yellow arrows indicate invasive tumors. d Weights of spleens. Data for b and d are mean ± SEM. * P < 0.05; ** P < 0.01; **** P < 0.0001; n.s.: not significant; by two-way ANOVA followed by Šídák's multiple comparisons test
PPARδ’s upregulation of Ccl20 chemoattracts, while GSK3787 inhibits, Ccr6+ immune cells infiltrating into gastric tissues of the mice
GAC development was strongly associated with chronic gastric inflammation in PpardTG mice [8], but the underlying mechanisms remained elusive. We previously reported that Ccl20 was the chemokine most markedly upregulated by transgenic PPARδ expression in VGPCs [8]. CCR6 is a non-promiscuous chemokine receptor that has only one known chemokine ligand, CCL20, and commonly exists on T cells and myeloid cells [19], the majority of gastric tumor–infiltrating immune cells in PpardTG mice [8]. To understand the biological significance and underlying mechanisms of PPARδ-mediated Ccl20 upregulation in the regulation of the gastric inflammation and GAC development, we performed in situ hybridization staining of RNAscope Duplex Assay for Ccl20 with villin, or with Ccr6 in stomachs and spleens of PpardTG mice and their WT littermates and found the following: (1) spleens of both PpardTG and WT mice had enriched Ccr6+ immune cells but no detectable Ccl20 mRNA expression (Fig. S1a). (2) Transgenic PPARδ overexpression in VGPCs significantly increased expansion and transformation of double Ccl20+ and villin+ VGPC cells and eventually drove these cells to form GAC as the PpardTG mice aged from 10 to 55 weeks (Fig. 3a, b). (3) As Ccl20 expression gradually increased in VGPCs, this transgenic PPARδ overexpression also gradually increased numbers of Ccr6+ immune cells infiltrating into the stomach, particularly into the gastric epithelial crypts or into gastric submucosa between the bottom of the gastric mucosa and the muscle layer in PpardTG mice from age 10 weeks to 55 weeks (Fig. 3c, d). CCR6 protein expression is significantly associated with metastasis, stage, and poor prognosis of GC [25]. We also retrieved and analyzed Stomach Adenocarcinoma (STAD) RNA-seq data from TCGA cancer database using The Human Protein Atlas’s Kaplan–Meier survival analysis tool and found that CCR6 mRNA expression was negatively associated with GC patient overall survival (Fig. 3e). In an in vitro study, Doxycycline (Dox)–induced human PPARδ overexpression in human GC cell lines AGS and N87 (Fig. S1b) and Dox–induced mouse PPARδ overexpression in mouse GC cells generated from GAC of 55-week-old PpardTG mice in our laboratory [8] (Fig. S1c) significantly increased CCL20 mRNA expression in these cells (Fig. 3f, g), indicating that PPARδ transcriptionally upregulated CCL20 in human and mouse GC cells. Ccr6+ and Ccr6− gastric tumor–infiltrating CD45+ immune cells from PpardTG mice were then sorted by flow cytometry. Ccr6+CD45+ immune cells had higher Ifng mRNA expression than Ccr6−CD45+ cells did (Fig. 3h), indicating that Ccl20 might chemoattract Ccr6+CD45+ cells via the Ccl20/Ccr6 axis into the gastric tissues to secrete Ifng, a strong pro-inflammation cytokine, to promote chronic inflammation and GAC development, which further supports our previous findings [8].
PPARδ upregulation of Ccl20 chemoattracted, but GSK3787 inhibited Ccr6+ immune cells infiltrating into gastric tissues. a–d Representative images of RNAscope Duplex Assay for Ccl20 and villin (a) and the quantitative scores of Ccl20 (b) or for Ccl20 and Ccr6 (c) and the quantitative scores of Ccr6 (d) in the stomachs of PpardTG and WT mice at the indicated ages. e Overall survival of GC patients with low vs. high CCR6 mRNA expression from analysis of Stomach Adenocarcinoma (STAD) RNA-seq data from TCGA cancer database. P value was calculated by Mantel-Cox test. f, g CCL20 mRNA expression was measured by RT-qPCR in AGS and N87 human GC (f) and mouse GC (g) cells transduced with human or mouse Dox-inducible PPARδ expression lentivirus with Doxycycline (2 µg/mL) or its dissolvent treatment for 48 h. h Ifng mRNA expression levels measured by RT-qPCR in flow cytometry–sorted Ccr6+ and Ccr6− stomach–infiltrating inflammatory immune cells from PpardTG mice at age 50 weeks. i–l The PpardTG mice and their WT littermates at 6–8 weeks were fed either a GSK3787 (200 mg/kg) or a control diet for 44 weeks and then euthanized as described in Fig. 2. i Gastric epithelial cells were scraped and examined for Ccl20 mRNA expression measured by RT-qPCR. j–l Representative images of in situ hybridization staining of RNAscope Duplex Assay for Ccl20 and Ccr6 (j) and the quantitative scores of Ccl20 (k) and Ccr6 (l) in stomachs of the indicated mice. Data are mean ± SEM for b, d, i, k and l. * P < 0.05; ** P < 0.01; *** P < 0.001; ****P < 0.0001; by one-way ANOVA followed by Tukey’s multiple comparisons test. Data are mean ± SD for f–h. ** P < 0.01; *** P < 0.001; by unpaired t test
In sharp contrast, GSK3787 significantly inhibited PPARδ–induced Ccl20 upregulation (Fig. 3i–k) and infiltration of Ccr6+ immune cells into stomachs in PpardTG mice (Fig. 3j, l) while suppressing GAC progression (Fig. 2). These data substantiate the critical role of PPARδ-Ccl20/Ccr6+ immune cell signaling alteration in chronic gastric inflammation and GAC development.
PPARδ promotes but GSK3787 inhibits gastric immune suppression in PpardTG mice
We next performed immune cell profiling to further characterize the subpopulations of the stomach-infiltrating immune cells that promote chronic gastric inflammation and GAC development. Stomachs of WT mice had very limited immune cells that were insufficient for further immune cell profiling [8]; thus, we performed multiple panels of immune cell profiling by flow cytometry on the gastric corpus tissues of PpardTG mice at age 50 weeks, when GAC developed, to define subsets of immune cells in the stomachs. The majority of stomach-infiltrating CD45+ immune cells from PpardTG mice were CD11b+ myeloid cells and CD3+ T cells (Fig. S2a). The immune cell phenotype of PpardTG tumors analyzed by FlowJo-tSNE (t-distributed stochastic neighbor embedding) showed more abundant tumor-associated macrophages (TAMs), polymorphonuclear (PMN)-MDSCs/neutrophils, monocytic (M)-MDSCs, and CD4+ T cells than CD8+ T cells and B cells (Fig. 4a [left]), as well as more abundant CCR6+ immune cells than CCR6− immune cells (Fig. 4a [right]). 32.86% ± 4.15% (mean ± SEM, hereafter unless otherwise specified), 14.60% ± 1.74%, and 31.04% ± 2.87% of stomach-infiltrating CD11b+ myeloid cells were TAMs (CD11b+F4/80+), M-MDSCs (CD11b+Ly6ChighLy6G−), and PMN-MDSCs/neutrophils (CD11b+Ly6ClowLy6G+), respectively (Fig. 4b, c). Not surprisingly, 68.80% ± 4.30% of CD45+ cells, 75.34% ± 5.99% of CD45+CD11b+ cells (Fig. S2b, c), 91.38% ± 2.49% of TAMs, 32.30% ± 4.64% of M-MDSCs and 44.33% ± 2.49% of PMN-MDSCs/neutrophils (Fig. 4d, e) in the stomachs of PpardTG mice were Ccr6+. Further analyses of CD3+ T subsets showed that the stomach-infiltrating CD3+ T cells had 1) 35.25% ± 0.21% CD4+ and 23.23% ± 0.72% T regulatory cells (Tregs, CD4+CD25+Foxp3+), indicating that Tregs were the majority of stomach infiltrating CD4+ T cells; 2) only 2.54% ± 0.77% CD8+ T cells, and 3) low ratios of CD8+ T cells to CD4+ T cells (mean ± SD: 0.07 [1:14 for CD8+: CD4+] ± 0.04) in PpardTG mice (Fig. 4f, g). The ratios of Ccr6+ cells of CD3+ (57.18% ± 9.01%) and CD4+ (61.48% ± 12.04%) were higher than those of CD8+ T cells (37.68% ± 8.17%) in the stomach of every examined mouse (Fig. 4h, i).
Subsets of total and Ccr6+ immune cells were profiled and quantified in stomachs of PpardTG mice. The PpardTG and WT mice at age 50 weeks were euthanized, and the stomachs were harvested and examined for immune cell profiling by flow cytometry. The WT mice had extremely low abundance of CD45+ immune cells in gastric tissues that precluded meaningful analyses. a Representative immune cell profiling by FlowJo-tSNE (t-distributed stochastic neighbor embedding) of stomach-infiltrating CD45+ cells based on parameters of CD45, CD11b, Ly6C, Ly6G, F4/80, CD3, CD4, CD8a and CCR6. The different colors indicated different immune cell subgroups, including macrophages, M-MDSCs, PMN-MDSCs/neutrophils, CD4+ T cells, CD8+ T cells, and B cells in PpardTG mice. b, c Representative flow cytometry images of stomach–infiltrating macrophages, M-MDSCs and PMN-MDSCs/neutrophils (b) and their percentages out of CD45+CD11b+ myeloid cells (c) in PpardTG mice (n = 5 mice). d, e Representative flow cytometry images (d) and ratios (e) of stomach–infiltrating Ccr6+ macrophages, Ccr6+ M-MDSCs, and Ccr6+ PMN-MDSCs/neutrophils in PpardTG mice (n = 4 mice). f, g Representative flow cytometry images (f) and percentages (g) of stomach–infiltrating CD4+ T, Tregs and CD8+ T out of CD3+ T cells in PpardTG mice. h, i Representative flow cytometry images (h) and ratios (i) of stomach– infiltrating Ccr6+ CD3+, Ccr6+CD4+ and Ccr6+CD8+ T cells in PpardTG mice. Data are mean ± SEM
Furthermore, the mechanistic studies with another method of immunohistochemistry staining showed that GSK3787 treatment significantly decreased F4/80+ macrophages (Fig. 5a [left], b), Ly6G+ MDSCs (Fig. 5a [right], c), and Foxp3+ Tregs (Fig. 5d [left], e), but increased CD8+ T cells (Fig. 5d [right], f) compared to control diet treatment in PpardTG mice. Taken together, our novel findings suggested that PPARδ promoted immune suppression through upregulating Ccl20 to recruit Ccr6+ immunosuppressive cells (e.g., TAMs, MDSCs, Tregs) into the stomachs; GSK3787 markedly reversed the PPARδ-induced immune suppression by decreasing TAMs, MDSCs and Tregs and increasing CD8+ T cells in the stomachs to suppress GAC progression in PpardTG mice.
GSK3787 inhibited PPARδ-mediated immune suppression in PpardTG mice. a–c Representative immunohistochemistry images (a) and quantitative data (b, c) for macrophages (F4/80+) and PMN-MDSCs/neutrophils (Ly6G+) in the stomachs of PpardTG mice fed a GSK3787 or control (Ctrl) diet as described in Fig. 2 (n = 5 mice per group). d–f Representative immunohistochemistry images (d) and quantitative data (e, f) for Tregs (Foxp3+) and CD8+ T cells in the mice as described in panel a–c. Data are mean ± SEM for b, c, e and f. ** P < 0.01; *** P < 0.001; by unpaired t test
Ccl20 is a potential circulating protein biomarker for the early detection and progression of GAC in PpardTG mice
It is critical to identify novel biomarkers for GAC early detection and progression to improve GAC patients’ outcomes. Our PpardTG GAC mouse model recapitulating human GAC pathogenesis provides us an ideal preclinical model to identify novel biomarkers. We therefore screened for a panel of chemokines in sera of PpardTG mice and sex- and age-matched WT littermates at ages of 10 weeks (before GC development), 25 weeks (at early stages of GC, i.e., hyperplasia and low-grade dysplasia) and 55 weeks (at late stages of GC, i.e., high-grade dysplasia and adenocarcinoma) [8] using the LEGENDplex Mouse Proinflammatory Chemokine Panel, which simultaneously quantifies 13 major inflammatory chemokines (Ccl2, Ccl3, Ccl4, Ccl5, Ccl11, Ccl17, Ccl20, Ccl22, Cxcl1, Cxcl5, Cxcl9, Cxcl10, and Cxcl13). Among these chemokines, Ccl20 levels were significantly higher in sera of PpardTG mice than in those of their WT littermates at age 10 weeks, before GAC development (Fig. 6a), suggesting its potential value as an early GAC detection marker. Furthermore, Ccl20 levels continuously increased as the PpardTG mice aged from 10 to 55 weeks while the tumor progressed (Fig. 6a), suggesting the potential value of Clc20 as a GAC progression marker. The Cxcl9 levels in sera were higher in PpardTG mice at ages 25 and 55 weeks than those in their WT littermates (Fig. 6b). In addition, Cxcl1, Cxcl5, Cxcl10, Cxcl13, and Ccl2-4 levels in sera were also significantly higher in PpardTG mice than in WT littermates at age 55 weeks (Fig. 6c–i). There was no difference in serum levels of Ccl5, Ccl11, Ccl22, or Ccl17 between PpardTG mice and WT littermates at all three examined ages (Fig. S3a–d).
A panel of 13 chemokines were measured and compared in sera of PpardTG mice and WT littermates without or with GSK3787 treatment. a–i The sera of PpardTG and WT littermates at 10, 25, and 55 weeks were collected and measured for a panel of 13 proinflammatory chemokines using the LEGENDplex Mouse Proinflammatory Chemokine Panel kit (n = 4–10 mice for age 10 weeks, n = 14 mice for age 25 weeks, and n = 26–28 mice for age 55 weeks per group). a–i The concentrations of Ccl20 (a), Cxcl9 (b), Cxcl1 (c), Cxcl5 (d), Cxcl10 (e), Cxcl13 (f), Ccl2 (g), Ccl3 (h) and Ccl4 (i) in the sera of the indicated mice. j, k The sera of PpardTG and WT littermates at 6–8 weeks fed a GSK3787 or control (Ctrl) diet for 44 weeks as described in Fig. 2 were collected and measured for the same panel of 13 proinflammatory chemokines as described in panel a–i (n = 6 per group). j The heatmap for three representative mice per group. Heatmap was generated by R 4.2.2 ‘pheatmap’ package. k The concentrations of Ccl20 in the sera of the indicated mice. Data are mean ± SEM for a–i, and k. * P < 0.05; ** P < 0.01; ***P < 0.001; ****P < 0.0001; n.s.: not significant; by two-way ANOVA followed by Šídák’s multiple comparisons test
Interestingly, GSK3787 treatment significantly decreased Ccl20 levels in sera of PpardTG mice (Fig. 6j, k) but had no effects on the other 12 measured chemokines (Fig. S4a–l), reinforcing a critical role of PPARδ in upregulation of Ccl20 during GC tumorigenesis. Our data indicated that Ccl20 is a potential circulating biomarker for the early detection, progression of GAC, and GAC treatment surveillance in the preclinical mouse model.
To further explore the clinical significance of these tested chemokines, we retrieved and analyzed the dataset GSE63089/GPL5175 using the NCBI GEO2R tool [ Casamayor M, Morlock R, Maeda H, Ajani J. Targeted literature review of the global burden of gastric cancer. Ecancermedicalscience. 2018;12:883. https://doi.org/10.3332/ecancer.2018.883. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660. Neels JG, Grimaldi PA. Physiological functions of peroxisome proliferator-activated receptor beta. Physiol Rev. 2014;94(3):795–858. https://doi.org/10.1152/physrev.00027.2013. Manickam R, Wahli W. Roles of peroxisome proliferator-activated receptor beta/delta in skeletal muscle physiology. Biochimie. 2017;136:42–8. https://doi.org/10.1016/j.biochi.2016.11.010. Wang D, Fu L, Ning W, Guo L, Sun X, Dey SK, et al. Peroxisome proliferator-activated receptor δ promotes colonic inflammation and tumor growth. Proc Natl Acad Sci U S A. 2014;111(19):7084–9. https://doi.org/10.1073/pnas.1324233111. Montagner A, Delgado MB, Tallichet-Blanc C, Chan JS, Sng MK, Mottaz H, et al. Src is activated by the nuclear receptor peroxisome proliferator-activated receptor β/δ in ultraviolet radiation-induced skin cancer. EMBO Mol Med. 2014;6(1):80–98. https://doi.org/10.1002/emmm.201302666. Xu M, Zuo X, Shureiqi I. Targeting peroxisome proliferator-activated receptor-beta/delta in colon cancer: how to aim? Biochem Pharmacol. 2013;85(5):607–11. https://doi.org/10.1016/j.bcp.2012.09.031. Zuo X, Deguchi Y, Xu W, Liu Y, Li HS, Wei D, et al. PPARD and interferon gamma promote transformation of gastric progenitor cells and tumorigenesis in mice. Gastroenterology. 2019. https://doi.org/10.1053/j.gastro.2019.03.018. Wagner N, Wagner KD. PPAR beta/delta and the hallmarks of cancer. Cells. 2020. https://doi.org/10.3390/cells9051133. Liu Y, Colby JK, Zuo X, Jaoude J, Wei D, Shureiqi I. The role of PPAR-δ in metabolism, inflammation, and cancer: many characters of a critical transcription factor. Int J Mol Sci. 2018. https://doi.org/10.3390/ijms19113339. Qiao XT, Ziel JW, McKimpson W, Madison BB, Todisco A, Merchant JL, et al. Prospective identification of a multilineage progenitor in murine stomach epithelium. Gastroenterology. 2007;133(6):1989–98. https://doi.org/10.1053/j.gastro.2007.09.031. Ma ES, Wang ZX, Zhu MQ, Zhao J. Immune evasion mechanisms and therapeutic strategies in gastric cancer. World J Gastrointest Oncol. 2022;14(1):216–29. https://doi.org/10.4251/wjgo.v14.i1.216. Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51(1):27–41. https://doi.org/10.1016/j.immuni.2019.06.025. Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27(8):1482–92. https://doi.org/10.1093/annonc/mdw168. Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell. 2019;35(4):559-72.e7. https://doi.org/10.1016/j.ccell.2019.02.008. Hou P, Kapoor A, Zhang Q, Li J, Wu CJ, Li J, et al. Tumor microenvironment remodeling enables bypass of oncogenic KRAS dependency in pancreatic cancer. Cancer Discov. 2020;10(7):1058–77. https://doi.org/10.1158/2159-8290.Cd-19-0597. Fan Y, Li Y, Yao X, ** J, Scott A, Liu B, et al. Epithelial SOX9 drives progression and metastases of gastric adenocarcinoma by promoting immunosuppressive tumour microenvironment. Gut. 2022. https://doi.org/10.1136/gutjnl-2021-326581. Kim W, Chu TH, Nienhüser H, Jiang Z, Del Portillo A, Remotti HE, et al. PD-1 signaling promotes tumor-infiltrating myeloid-derived suppressor cells and gastric tumorigenesis in mice. Gastroenterology. 2021;160(3):781–96. https://doi.org/10.1053/j.gastro.2020.10.036. Lee AY, Eri R, Lyons AB, Grimm MC, Korner H. CC chemokine ligand 20 and its cognate receptor CCR6 in mucosal T cell immunology and inflammatory bowel disease: odd couple or axis of evil? Front Immunol. 2013;4:194. https://doi.org/10.3389/fimmu.2013.00194. Kadomoto S, Izumi K, Mizokami A. The CCL20-CCR6 axis in cancer progression. Int J Mol Sci. 2020. https://doi.org/10.3390/ijms21155186. Chen W, Qin Y, Liu S. CCL20 signaling in the tumor microenvironment. Adv Exp Med Biol. 2020;1231:53–65. https://doi.org/10.1007/978-3-030-36667-4_6. Zuo X, Xu M, Yu J, Wu Y, Moussalli MJ, Manyam GC, et al. Potentiation of colon cancer susceptibility in mice by colonic epithelial PPAR-delta/beta overexpression. J Natl Cancer Inst. 2014;106(4):dju052. https://doi.org/10.1093/jnci/dju052. Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–33. https://doi.org/10.18632/oncotarget.10337. Palkar PS, Borland MG, Naruhn S, Ferry CH, Lee C, Sk UH, et al. Cellular and pharmacological selectivity of the peroxisome proliferator-activated receptor-beta/delta antagonist GSK3787. Mol Pharmacol. 2010;78(3):419–30. https://doi.org/10.1124/mol.110.065508. Han G, Wu D, Yang Y, Li Z, Zhang J, Li C. CrkL meditates CCL20/CCR6-induced EMT in gastric cancer. Cytokine. 2015;76(2):163–9. https://doi.org/10.1016/j.cyto.2015.05.009. Zhang X, Ni Z, Duan Z, **n Z, Wang H, Tan J, et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS ONE. 2015;10(2):e0116979. https://doi.org/10.1371/journal.pone.0116979. Pollock CB, Rodriguez O, Martin PL, Albanese C, Li X, Kopelovich L, et al. Induction of metastatic gastric cancer by peroxisome proliferator-activated receptordelta activation. PPAR Res. 2010;2010:571783. https://doi.org/10.1155/2010/571783. Song S, Wang Z, Li Y, Ma L, ** J, Scott AW, et al. PPARδ interacts with the hippo coactivator YAP1 to promote SOX9 expression and gastric cancer progression. Mol Cancer Res MCR. 2020;18(3):390–402. https://doi.org/10.1158/1541-7786.Mcr-19-0895. Vogiatzi P, Cassone M, Luzzi I, Lucchetti C, Otvos L Jr, Giordano A. Helicobacter pylori as a class I carcinogen: physiopathology and management strategies. J Cell Biochem. 2007;102(2):264–73. https://doi.org/10.1002/jcb.21375. Palrasu M, Zaika E, El-Rifai W, Garcia-Buitrago M, Piazuelo MB, Wilson KT, et al. Bacterial CagA protein compromises tumor suppressor mechanisms in gastric epithelial cells. J Clin Invest. 2020;130(5):2422–34. https://doi.org/10.1172/jci130015. Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–9. https://doi.org/10.1056/NEJMoa001999. Mera RM, Bravo LE, Camargo MC, Bravo JC, Delgado AG, Romero-Gallo J, et al. Dynamics of Helicobacter pylori infection as a determinant of progression of gastric precancerous lesions: 16-year follow-up of an eradication trial. Gut. 2018;67(7):1239–46. https://doi.org/10.1136/gutjnl-2016-311685. Khatoon J, Rai RP, Prasad KN. Role of Helicobacter pylori in gastric cancer: updates. World J Gastrointest Oncol. 2016;8(2):147–58. https://doi.org/10.4251/wjgo.v8.i2.147. Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, et al. Helicobacter pylori activates and expands Lgr5(+) stem cells through direct colonization of the gastric glands. Gastroenterology. 2015;148(7):1392-404.e21. https://doi.org/10.1053/j.gastro.2015.02.049. Liabeuf D, Oshima M, Stange DE, Sigal M. Stem Cells, Helicobacter pylori, and mutational landscape: utility of preclinical models to understand carcinogenesis and to direct management of gastric cancer. Gastroenterology. 2022;162(4):1067–87. https://doi.org/10.1053/j.gastro.2021.12.252. Nagy TA, Wroblewski LE, Wang D, Piazuelo MB, Delgado A, Romero-Gallo J, et al. beta-Catenin and p120 mediate PPARdelta-dependent proliferation induced by Helicobacter pylori in human and rodent epithelia. Gastroenterology. 2011;141(2):553–64. https://doi.org/10.1053/j.gastro.2011.05.004. Wang K, Shen T, Siegal GP, Wei S. The CD4/CD8 ratio of tumor-infiltrating lymphocytes at the tumor-host interface has prognostic value in triple-negative breast cancer. Hum Pathol. 2017;69:110–7. https://doi.org/10.1016/j.humpath.2017.09.012. Zurlo IV, Schino M, Strippoli A, Calegari MA, Cocomazzi A, Cassano A, et al. Predictive value of NLR, TILs (CD4+/CD8+) and PD-L1 expression for prognosis and response to preoperative chemotherapy in gastric cancer. Cancer Immunol Immunother. 2022;71(1):45–55. https://doi.org/10.1007/s00262-021-02960-1. Lee PH, Yang TY, Chen KC, Huang YH, Tseng JS, Hsu KH, et al. Higher CD4/CD8 ratio of pleural effusion predicts better survival for lung cancer patients receiving immune checkpoint inhibitors. Sci Rep. 2021;11(1):9381. https://doi.org/10.1038/s41598-021-89043-4. Liu Y, Deguchi Y, Wei D, Liu F, Moussalli MJ, Deguchi E, et al. Rapid acceleration of KRAS-mutant pancreatic carcinogenesis via remodeling of tumor immune microenvironment by PPARδ. Nat Commun. 2022;13(1):2665. https://doi.org/10.1038/s41467-022-30392-7. Abrego J, Sanford-Crane H, Oon C, **ao X, Betts CB, Sun D, et al. A cancer cell-intrinsic GOT2-PPARδ axis suppresses antitumor immunity. Cancer Discov. 2022;12(10):2414–33. https://doi.org/10.1158/2159-8290.Cd-22-0661. Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, et al. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25(17):2029–44. https://doi.org/10.3748/wjg.v25.i17.2029. Camargo MC, Song M, Sawada N, Inoue M, Shimazu T, Charvat H, et al. Prediagnostic circulating inflammation-related biomarkers and gastric cancer: a case-cohort study in Japan. Cytokine. 2021;144:155558. https://doi.org/10.1016/j.cyto.2021.155558. Shen Q, Polom K, Williams C, de Oliveira FMS, Guergova-Kuras M, Lisacek F, et al. A targeted proteomics approach reveals a serum protein signature as diagnostic biomarker for resectable gastric cancer. EBioMedicine. 2019;44:322–33. https://doi.org/10.1016/j.ebiom.2019.05.044. Song M, Rabkin CS, Torres J, Kemp TJ, Zabaleta J, Pinto LA, et al. Circulating inflammation-related markers and advanced gastric premalignant lesions. J Gastroenterol Hepatol. 2019;34(5):852–6. https://doi.org/10.1111/jgh.14518.References
Acknowledgements
This work was supported by the National Cancer Institute grants (R01CA236905 and R03CA235106 to X. Z; R01CA142969 and R01CA266223 to I.S.); the Cancer Prevention and Research Institute of Texas grants (RP140224 to I.S.); the University of Texas MD Anderson internal research grant (PS103703 to X.Z); Texas Medical Center Digestive Diseases Center seed grant (Record#: AWD00005436 by sponsor award 5P30DK056338-18 to X.Z); the University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment (#123264 to D.W. ), and the University of Texas MD Anderson Cancer Center Institutional Research Program (#125092 to D.W). This study made use of the MD Anderson Cancer Center Genetically Engineered Mouse Facility and Flow Cytometry and Cellular Imaging Facility and was supported by Cancer Center Support Grant P30CA016672. The manuscript was edited by Sarah Bronson, ELS, of the Research Medical Library at The University of Texas MD Anderson Cancer Center.