Abstract
Some recent papers clearly indicate that the cytoplasmic domain of KcsA plays a role in pH sensing. We have performed, for the first time, a targeted molecular dynamics (TMD) simulation of the opening of full-length KcsA at pH 4 and pH 7, with a special interest for the cytoplasmic domain. Association energy calculations show a stabilization at pH 7 confirming that the protonation of some amino-acids at pH 4 in this domain plays a role in the opening process. A careful analysis of the pH dependent charges borne by residues in the cytoplasmic domain and their interactions confirms some literature experimental data and permits to give further insight into the role played by some of them in the opening process.











Similar content being viewed by others
Notes
Except for the full systems simulations where residues 113 and 114 were not restrained since they are located at the border between TM and cytoplasmic domains.
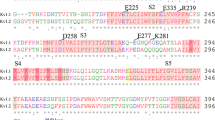
The global bending of the cytoplasmic domain was defined by the angle between the mass-weighted backbone atoms of residues Val48, Ala111 and Asp120 of the four units.
References
Hille B (2003) Ion channels of excitable membranes, 3rd edn. Sinauer, Sunderland, MA
Alagem N, Yesylevskyy S, Reuveny E (2003) The pore helix is involved in stabilizing the open state of inwardly rectifying K + channels. Biophys J 85:300–312
Krol E, Trebacz K (2000) Ways of ion channel gating in plant cells. Ann Bot 86:449–469
Cuello LG, Cortes DM, Jogini V, Sompornpisut A, Perozo E (2010) A molecular mechanism for proton-dependent gating in KCSA. FEBS Lett 584:1126–1132
Salkoff L, Wei AD, Baban B, Butler A, Fawcett G, Ferreira G, Santi CM (2005) The C. elegans research community, WormBook, doi:10.1895/wormbook.1.42.1
Uysal S, Vasquez V, Terechko V, Esaki K, Koide S, Fellouse FA, Sidhu SS, Perozo E, Kossiakoff A (2009) The crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci USA 106:6644–6649
Cuello LG, Romero JG, Cortes DM, Perozo E (1998) pH-dependent gating in the Streptomyces lividans K channel. Biochemistry 37:3229–3236
Heginbotham L, LeMasurier M, Kolmakova-Partensky L, Miller C (1999) Single streptomyces lividans K + channels: functional asymmetries and sidedness of proton activation. J Gen Physiol 114:551–560
Schrempf H, Schmidt O, Kümmerlen R, Hinnah S, Müller D, Betzler M, Steinkamp T, Wagner R (1995) A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J 14:5170–5178
Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R (1998) The structure of the potassium channel: molecular basis of K + conduction and selectivity. Science 280:69–77
Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414:43–48
Pau VP, Zhu Y, Yuchi Z, Hoang QQ, Yang DS (2007) Characterization of the C-terminal domain of a potassium channel from Streptomyces lividans (KcsA). J Biol Chem 282:29163–29169
Cortes DM, Cuello LG, Perozo E (2001) Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol 117:165–180
Perozo E, Cortes DM, Cuello LG (1999) Structural rearrangements underlying K1-channel activation gating. Science 285:73–78
Liu YS, Sompornpisut P, Perozo E (2001) Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol 8:883–887
Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R (2002) The open pore conformation of potassium channels. Nature 417:523–526
Thompson AN, Posson DJ, Parsa PV, Nimigean CM (2008) Molecular mechanism of pH sensing in KcsA potassium channels. Proc Natl Acad Sci USA 105:6900–6905
Takeuchi K, Takahashi H, Kawano S, Shimada I (2007) Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem 282:15179–15186
Shen Y, Kong Y, Ma J (2002) Intrinsic flexibility and gating mechanism of the potassium channel KcsA. Proc Natl Acad Sci USA 99:1949–1953
Hirano M, Takeuchi Y, Aoki T, Yanagida T, Ide T (2010) Rearrangements in the KcsA cytoplasmic domain underlie its gating. J Biol Chem 285:3777–3783
Uysal S, Cuello LG, Cortes DM, Koide S, Kossiakoff AA, Perozo E (2011) Mechanism of activation gating in the full-length KcsA K + channel. Proc Natl Acad Sci USA 108:11896–11899
Zhong W, Guo W (2009) Mixed modes in opening of KcsA potassium channel from a targeted molecular dynamics simulation. Biochem Biophys Res Comm 388:86–90
Compoint M, Picaud F, Ramseyer C, Girardet C (2005) Zip gating of the KcsA channel studied by targeted molecular dynamics. Chem Phys Lett 407:199–204
Compoint M, Picaud F, Ramseyer C, Girardet C (2005) Targeted molecular dynamics of an open-state KcsA channel. J Chem Phys 122:134707–134714
Zhong W, Guo W, Ma S (2008) Intrinsic aqueduct orifices facilitate K + channel gating. FEBS Lett 582:3320–3324
Holyoake J, Domene C, Bright JN, Sansom MS (2004) KcsA closed and open: modelling and simulation studies. Eur Biophys J 33:238–246
Cuello LG, Jogini V, Cortes DM, Perozo E (2010) Structural mechanism of C-type inactivation in K+ channels. Nature 466:203–208
Compoint M, Carloni P, Ramseyer C, Girardet C (2004) Molecular dynamics study of the KcsA channel at 2.0 Å resolution: Stability and concerted motions within the pore. Biochim Biophys.Acta, 1661:26–39
Morais-Cabral JH, Zhou Y, MacKinnon R (2001) Energetic optimization of ion conduction rate by the K + selectivity filter. Nature 414:37–42
Raja M (2010) The role of extramembranous cytoplasmic termini in assembly and stability of the tetrameric K(+)-channel KcsA. J Membrane Biol 235:51–61
Heginbotham L, Odessey E, Miller C (1997) Tetrameric stoichiometry of a prokaryotic K + channel. Biochemistry 36:10335–10342
Meuser D, Splitt H, Wagner R, Schrempf H (1999) Exploring the open pore of the potassium channel from Streptomyces lividans. FEBS Lett 462:447–452
Hirano M, Onishi Y, Yanagida T, Ide T (2011) Role of the KcsA channel cytoplasmic domain in pH-dependent gating. Biophys J 101:2157–2162
Cuello LG, Jogini V, Cortes DM, Sompornpisut A, Purdy MD, Wiener MC, Perozo E (2010) Design and characterization of a constitutively open KcsA. FEBS Lett 584:1133–1138
Cuello LG, Jogini V, Cortes DM, Pan AC, Gagnon DG, Dalmas O, Cordero-Morales JF, Chakrapani S, Roux B, Perozo E (2010) Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature 466:272–275
Kharkyanen VN, Yesylevskyy SO, Berezetskaya NM, Boiteux C, Ramseyer C (2009) Semi-quantitative model of the gating of KcsA ion channel. 2. Dynamic self-organization model of the gating Biopolym. Cell 25:476–483
Andersen HC (1980) Molecular dynamics at constant pressure and/or temperature. J Chem Phys 72:2384–2393
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 33:W368–W371
Schlitter I, Engels M, Krüger P (1994) Targeted molecular dynamics: a new approach for searching pathways of conformational transition. J Mol Graphics 12:84–89
Karplus M, Kuriyan J (2005) Molecular dynamics and protein function. Proc Natl Acad Sci USA 102:6679–6685
Case D, Darden T, Cheatham TE, Simmerling C, Wang J, Duke, Luo R, Crowley M, Walker R, Zhang W, Merz K, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong K, Paesani F, Vanicek J, Wu X, Brozell S, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Gui G, Mathews D, Seetin M, Sagui C, Babin V, Kollman P (2010) AMBER 11, University of California, San Francisco
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of N-alkanes. J Comput Phys 23:327–341
Kottalam J, Case D (1990) Langevin modes of macromolecules: applications to crambin and DNA hexamers. Biopolymers 29:1409–1421
Jörg W, Peter SS, Still WC (1999) Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J Comput Chem 20:217–230
Roberts E, Eargle J, Wright D, Luthey-Schulten Z (2006) Multiseq: unifying sequence and structure data for evolutionary analysis. BMC Bioinforma 7:382–393
Chen X, Poon BK, Dousis A, Wang Q, Ma J (2007) Normal-mode refinement of anisotropic thermal parameters for potassium channel KcsA at 3.2 Å crystallographic resolution. Structure 15:955–962
Acknowledgments
The China Scholarship Council is gratefully acknowledged for granting a PhD scholarship to Yan LI.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 419 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Barbault, F., Delamar, M. et al. Targeted molecular dynamics (TMD) of the full-length KcsA potassium channel: on the role of the cytoplasmic domain in the opening process. J Mol Model 19, 1651–1666 (2013). https://doi.org/10.1007/s00894-012-1726-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1726-3