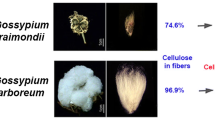
Abstract
Cellulose synthases (CesAs) are multi-subunit enzymes found on the plasma membrane of plant cells and play a pivotal role in cellulose production. The cotton fiber is mainly composed of cellulose, and the genetic relationships between CesA genes and cotton fiber yield and quality are not fully understood. Through a phylogenetic analysis, the CesA gene family in diploid Gossypium arboreum and Gossypium raimondii, as well as tetraploid Gossypium hirsutum (‘TM-1’) and Gossypium barbadense (‘Hai-7124’ and ‘3-79’), was divided into 6 groups and 15 sub-groups, with each group containing two to five homologous genes. Most CesA genes in the four species are highly collinear. Among the five cotton genomes, 440 and 1929 single nucleotide polymorphisms (SNPs) in the CesA gene family were identified in exons and introns, respectively, including 174 SNPs resulting in amino acid changes. In total, 484 homeologous SNPs between the A and D genomes were identified in diploids, while 142 SNPs were detected between the two tetraploids, with 32 and 82 SNPs existing within G. hirsutum and G. barbadense, respectively. Additionally, 74 quantitative trait loci near 18 GhCesA genes were associated with fiber quality. One to four GhCesA genes were differentially expressed (DE) in ovules at 0 and 3 days post anthesis (DPA) between two backcross inbred lines having different fiber lengths, but no DE genes were identified between these lines in develo** fibers at 10 DPA. Twenty-seven SNPs in above DE CesA genes were detected among seven cotton lines, including one SNP in Ghi_A08G03061 that was detected in four G. hirsutum genotypes. This study provides the first comprehensive characterization of the cotton CesA gene family, which may play important roles in determining cotton fiber quality.




Similar content being viewed by others
Abbreviations
- SNP:
-
Single nucleotide polymorphisms
- QTL:
-
Quantitative trait loci
- SNP:
-
Single nucleotide polymorphism
- DPA:
-
Day post-anthesis
- CesAs :
-
Cellulose synthase genes
- MAS:
-
Marker-assisted selection
- Gb:
-
Gossypium barbadense.
- Gh:
-
Gossypium hirsutum L.
References
Abdurakhmonov IY, Kohel RJ, Yu JZ, Pepper AE, Abdullaev AA, Kushanov FN, Salakhutdinov IB, Buriev ZT, Saha S, Scheffler BE, Jenkins JN, Abdukarimov A (2008) Molecular diversity and association map** of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics 92:478–487
Basra AS, Malik CP (1984) Development of the cotton fiber. Int Rev Cytol 89:65–113
Boli Y, Hu W, Li X, Cao J, Fan L (2015) Over-expression of GhUGP1 in Upland cotton improves fibre quality and reduces fibre sugar content. Plant Breed 134:197–202
Cao S, Cheng H, Zhang J, Aslam M, Yan M, Hu A, Lin L, Ojolo SP, Zhao H, Priyadarshani S, Yu Y, Cao G, Qin Y (2019) Genome-wide identification, expression pattern analysis and evolution of the Ces/Csl Gene superfamily in Pineapple (Ananas comosus). Plants (Basel) 8:275
Doblin M, Melis L, Newbigin E, Bacic A, Read S (2001) Pollen tubes of Nicotiana alata express two genes from different beta-glucan synthase families. Plant Physiol 125:2040–2052
Fang L, Wang Q, Hu Y, Jia Y, Chen J, Liu B, Zhang Z, Guan X, Chen S, Zhou B, Mei G, Sun J, Pan Z, He S, **ao S, Shi W, Gong W, Liu J, Ma J, Cai C, Zhu X, Guo W, Du X, Zhang T (2017a) Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet 49:1089–1098
Fang XM, Liu XY, Wang XQ, Wang WW, Liu DX, Zhang J, Liu DJ, Teng ZH, Tan ZY, Liu F, Zhang FJ, Jiang MC, Jia XL, Zhong JW, Yang JH, Zhang ZS (2017b) Fine-map** qFS07.1 controlling fiber strength in upland cotton (Gossypium hirsutum L.). Theor Appl Genet 130(4):795–806
Gapare W, Conaty W, Zhu QH, Liu S, Stiller W, Llewellyn D, Wilson I (2017) Genome-wide association study of yield components and fibre quality traits in a cotton germplasm diversity panel. Euphytica 213(3):66
Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin T, Persson S, Somerville C (2010) Identification of a novel CESA-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA 107:12866–12871
Han Z, Qin Y, Li X, Yu J, Li R, **ng C, Song M, Wu J, Zhang J (2020) A genome-wide analysis of pentatricopeptide repeat (PPR) protein-encoding genes in four Gossypium species with an emphasis on their expression in floral buds, ovules, and fibers in upland cotton. Mol Genet Genomics 295:55–66
Hu H, Zhang R, Tao Z, Li X, Li Y, Huang J, Li X, Han X, Feng S, Zhang G, Peng L (2018b) Cellulose synthase mutants distinctively affect cell growth and cell wall integrity for plant biomass production in Arabidopsis. Plant Cell Physiol 59:1144–1157
Hu H, Zhang R, Feng S, Wang Y, Wang Y, Fan C, Li Y, Liu Z, Schneider R, **a T, Ding S, Persson S, Peng L (2018a) Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol J 16:976–988
Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, Baruch K, Fang D, Liu X, Ruan YL, Rahman MU, Han J, Wang K, Wang Q, Wu H, Mei G, Zang Y, Han Z, Xu C, Shen W, Yang D, Si Z, Dai F, Zou L, Huang F, Bai Y, Zhang Y, Brodt A, Ben-Hamo H, Zhu X, Zhou B, Guan X, Zhu S, Chen X, Zhang T (2019) Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51:739–748
Huang G, Wu Z, Percy RG, Bai M, Li Y, Frelichowski J, Hu J, Wang K, Yu J, Zhu Y (2020) Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat Genet 52:516–524
Islam M, Zeng L, Thyssen G, Delhom C, Fang D (2016) Map** by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor Appl Genet 129:1071–1086
Jamshed M, Jia F, Gong J, Palanga KK, Shi Y, Li J, Shang H, Liu A, Chen T, Zhang Z, Cai J, Ge Q, Liu Z, Lu Q, Deng X, Tan Y, Or Rashid H, Sarfraz Z, Hassan M, Gong W, Yuan Y (2016) Identification of stable quantitative trait loci (QTLs) for fiber quality traits across multiple environments in Gossypium hirsutum recombinant inbred line population. BMC Genomics 17:197
Kumar M, Thammannagowda S, Bulone V, Chiang V, Han KH, Joshi CP, Mansfield SD, Mellerowicz E, Sundberg B, Teeri T, Ellis BE (2009) An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci 14(5):248–254
Lacape JM, Llewellyn D, Jacobs J, Arioli T, Becker D, Calhoun S, Al-Ghazi Y, Liu S, Palai O, Georges S, Giband M, de Assuncao H, Barroso PA, Claverie M, Gawryziak G, Jean J, Vialle M, Viot C (2010) Meta-analysis of cotton fiber quality QTLs across diverse environments in a Gossypiumhirsutum x G.barbadense RIL population. BMC Plant Biol 10:132
Lee JJ, Woodward AW, Chen ZJ (2007) Gene expression changes and early events in cotton fibre development. Ann Bot 100:1391–1401
Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, Liu J, Warburton ML, Cheng Y, Hao X, Zhang P, Zhao J, Liu Y, Wang G, Li J, Yan J (2013b) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45:43–50
Li A, **a T, Xu W, Chen T, Li X, Fan J, Wang R, Feng S, Wang Y, Wang B, Peng L (2013a) An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense. Planta 237:1585–1597
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, **ao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46:567–572
Li F, Fan G, Lu C, **ao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, Liang X, Huang G, Percy RG, Liu K, Yang W, Chen W, Du X, Shi C, Yuan Y, Ye W, Liu X, Zhang X, Liu W, Wei H, Wei S, Huang G, Zhang X, Zhu S, Zhang H, Sun F, Wang X, Liang J, Wang J, He Q, Huang L, Wang J, Cui J, Song G, Wang K, Xu X, Yu JZ, Zhu Y, Yu S (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33:524–530
Li W, Shang H, Ge Q, Zou C, Cai J, Wang D, Fan S, Zhang Z, Deng X, Tan Y, Song W, Li P, Koffi PK, Jamshed M, Lu Q, Gong W, Li J, Shi Y, Chen T, Gong J, Liu A, Yuan Y (2016b) Genome-wide identification, phylogeny, and expression analysis of pectin methylesterases reveal their major role in cotton fiber development. BMC Genomics 17:1000
Li S, Bashline L, Zheng Y, **n X, Huang S, Kong Z, Kim S, Cosgrove D, Gu Y (2016a) Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc Natl Acad Sci U S A 113:11348–11353
Li X, Wu M, Liu G, Pei W, Zhai H, Yu J, Zhang J, Yu S (2017) Identification of candidate genes for fiber length quantitative trait loci through RNA-Seq and linkage and physical map** in cotton. BMC Genomics 18:427
Li G, Liu X, Liang Y, Zhang Y, Cheng X, Cai Y (2020) Genome-wide characterization of the cellulose synthase gene superfamily in Pyrus bretschneideri and reveal its potential role in stone cell formation. Funct Integr Genomics 20(5):723–738
Lin Z, Wang Y, Zhang X, Zhang J (2012) Functional markers for cellulose synthase and their comparison to SSRs in cotton. Plant Mol Biol Rep 30:1270–1275
Liu X, Zhao B, Zheng HJ, Hu Y, Lu G, Yang CQ, Chen JD, Chen JJ, Chen DY, Zhang L, Zhou Y, Wang LJ, Guo WZ, Bai YL, Ruan JX, Shangguan XX, Mao YB, Shan CM, Jiang JP, Zhu YQ, ** L, Kang H, Chen ST, He XL, Wang R, Wang YZ, Chen J, Wang LJ, Yu ST, Wang BY, Wei J, Song SC, Lu XY, Gao ZC, Gu WY, Deng X, Ma D, Wang S, Liang WH, Fang L, Cai CP, Zhu XF, Zhou BL, Jeffrey Chen Z, Xu SH, Zhang YG, Wang SY, Zhang TZ, Zhao GP, Chen XY (2015) Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci Rep 5:14139
Ma Q, Wu M, Pei W, Wang X, Zhai H, Wang W, Li X, Zhang J, Yu J, Yu S (2016) RNA-seq-mediated transcriptome analysis of a fiberless mutant cotton and its possible origin based on SNP markers. PLoS ONE 11:e0151994
Mazarei M, Baxter HL, Li M, Biswal AK, Kim K, Meng X, Pu Y, Wuddineh WA, Zhang JY, Turner GB, Sykes RW, Davis MF, Udvardi MK, Wang ZY, Mohnen D, Ragauskas AJ, Labbé N, NealStewart JC (2018) Functional analysis of cellulose synthase CesA4 and CesA6genes in Switchgrass (Panicum virgatum) by overexpression and RNAi-mediated gene silencing. Front Plant Sci 9:1114
McFarlane H, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65(1):69–94
Nie X, Huang C, You C, Li W, Zhao W, Shen C, Zhang B, Wang H, Yan Z, Dai B, Wang M, Zhang X, Lin Z (2016) Genome-wide SSR-based association map** for fiber quality in nation-wide upland cotton inbreed cultivars in China. BMC Genomics 17:352
Ning Z, Chen H, Mei H, Zhang T (2014) Molecular tagging of QTLs for fiber quality and yield in the upland cotton cultivar Acala-Prema. Euphytica 195:143–156
Osako Y, Takata N, Nuoendagula IS, Umezawa T, Taniguchi T, Kajita S (2019) Expression analysis of cellulose synthases that comprise the Type II complex in hybrid aspen. Plant Biol 21:361–370
Paterson AH, Saranga Y, Menz M, Jiang CX, Wright RJ (2003) QTL analysis of genotype x environment interactions affecting cotton fiber quality. Theor Appl Genet 106:384–396
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, ** D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Pear JR, Kawagoet Y, Schreckengost WE, Delmert DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci U S A 93:12637–12642
Said J, Lin Z, Zhang X, Song M, Zhang J (2013) Comprehensive meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genomics 14:776
Said J, Knapka J, Song M, Zhang J (2015a) Cotton QTLdb: a cotton QTL database for QTL analysis, visualization, and comparison between Gossypiumhirsutum and G.hirsutum x G.barbadense populations. Mol Genetic Genomics 290(4):1615–1625
Said J, Song M, Wang H, Lin Z, Zhang X, Fang D, Zhang J (2015b) A comparative Meta-analysis of QTL between intraspecific Gossypiumhirsutum and interspecific G.hirsutum x G.barbadense populations. Mol Genetic Genomics 290(3):1003–1025
Sariful IM, Thyssen GN, Jenkins JN, Linghe Z, Delhom CD, Mccarty JC, Deng DD, Hinchliffe DJ, Jones DC, Fang DD (2016) A MAGIC population-based genome-wide association study reveals functional association of GhRBB1_A07 gene with superior fiber quality in cotton. BMC Genomics 17(1):903
Saxena IM, Brown MR (2008) Biochemistry and molecular biology of cellulose biosynthesis in plants: prospects for genetic engineering. Adv Plant Biochem Mol Biol 1:135–160
Shao Q, Zhang F, Tang S, Liu Y, Fang X, Liu D, Liu D, Zhang J, Teng Z, Paterson AH, Zhang Z (2014) Identifying QTL for fiber quality traits with three upland cotton (Gossypium hirsutum L.) populations. Euphytica 198:43–58
Song X, Xu L, Yu J, Tian P, Hu X, Wang Q, Pan Y (2018) Genome-wide characterization of the cellulose synthase gene superfamily in Solanum lycopersicum. Gene 688:71–83
Sun Z, Wang X, Liu Z, Gu Q, Zhang Y, Li Z, Ke H, Yang J, Wu J, Wu L, Zhang G, Zhang C, Ma Z (2017) Genome-wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol J 15(8):982–996
Udall JA, Long E, Hanson C, Yuan D, Ramaraj T, Conover J, Gong L, Arick M, Grover C, Peterson D, Wendel J (2019) De novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 (Bethesda) 9(10):3079–3085
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44:1098–1103
Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G, Ye Z, Huang H, Yan F, Ma Y, Zhang L, Liu M, You J, Yang Y, Liu Z, Huang F, Li B, Qiu P, Zhang Q, Zhu L, ** S, Yang X, Min L, Li G, Chen LL, Zheng H, Lindsey K, Lin Z, Udall JA, Zhang X (2019) Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet 51:224–229
Wu J, Zhang M, Zhang B, Zhang X, Guo L, Qi T, Wang H, Zhang J, **ng C (2017) Genome-wide comparative transcriptome analysis of CMS-D2 and its maintainer and restorer lines in upland cotton. BMC Genomics 18:454
Yang X, Zhou X, Wang X, Li Z, Zhang Y, Liu H, Wu L, Zhang G, Yan G, Ma Z (2015) Map** QTL for cotton fiber quality traits using simple sequence repeat markers, conserved intron-scanning primers, and transcript-derived fragments. Euphytica 201:215–230
Yin Y, Johns MA, Cao H, Rupani M (2014) A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genomics 15:260
Yuan D, Tang Z, Wang M, Gao W, Tu L, ** X, Chen L, He Y, Zhang L, Zhu L, Li Y, Liang Q, Lin Z, Yang X, Liu N, ** S, Lei Y, Ding Y, Li G, Ruan X, Ruan Y, Zhang X (2015) The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep 5:17662
Zhang JF, Stewart JM (2000) Economical and rapid method for extracting cotton genomic DNA. J Cotton Sci 4:193–201
Zhang Y, Wang XF, Li ZK, Zhang GY, Ma ZY (2011) Assessing genetic diversity of cotton cultivars using genomic and newly developed expressed sequence tag-derived microsatellite markers. Genet Mol Res 10:1462–1470
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, **ang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537
Zou X, Zhen Z, Ge Q, Fan S, Liu A, Gong W, Li J, Gong J, Shi Y, Wang Y, Liu R, Duan L, Lei K, Zhang Q, Jiang X, Zhang S, Jia T, Zhang L, Shang H, Yuan Y (2018) Genome-wide identification and analysis of the evolution and expression patterns of the cellulose synthase gene superfamily in Gossypium species. Gene 646:28–38
Acknowledgments
We thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of an early draft of this manuscript.
Funding
This work was supported by New Mexico Agricultural Experiment Station and a USDA-Hatch Project. It was financially supported by Cotton Heterosis Utilization Technology and Superior Hybrid Seed Creation Project (2016YFD0101415), the Hebei Province "Giant Plan" Innovation and Entrepreneurship Team, and the State Key Laboratory of Cotton Biology Open Fund (CB2018A15).
Author information
Authors and Affiliations
Contributions
SZ performed the study and drafted the manuscript. ZJ shared the work of verifying the existence of SNP. JC and ZH helped in screening of differential sites and software use, JC helped ZJ in the verifying the existence of SNP. XL was was responsible for RNA-Seq data. JY and CX collected flowering buds, extracted RNA and analyzed RNA-Seq data. MS searched the QTLs near the target genes, JW helped to add detailed information on RNA-seq experiment, FL assisted MS to find the physical location of the QTLs, JZ was responsible for screening representative materials for sequencing to verify the authenticity of SNPs. XZ helped to conceive the project. JZ conceived and oversaw the project and revised the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
438_2020_1758_MOESM5_ESM.xlsx
Sequence variations in the CesA gene family among homologous genes in different species identified in the NCBI databases (XLSX 35 KB)
438_2020_1758_MOESM6_ESM.xlsx
Detection of SNPs located in the coding sequences of five CesA genes in seven cotton lines using Sanger sequencing (XLSX 14 KB)
Rights and permissions
About this article
Cite this article
Zhang, S., Jiang, Z., Chen, J. et al. The cellulose synthase (CesA) gene family in four Gossypium species: phylogenetics, sequence variation and gene expression in relation to fiber quality in Upland cotton. Mol Genet Genomics 296, 355–368 (2021). https://doi.org/10.1007/s00438-020-01758-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01758-7