Abstract
Main conclusion
Molecular mechanisms of biological rhythms provide opportunities to harness functional allelic diversity in core (and trait- or stress-responsive) oscillator networks to develop more climate-resilient and productive germplasm.
Abstract
The circadian clock senses light and temperature in day–night cycles to drive biological rhythms. The clock integrates endogenous signals and exogenous stimuli to coordinate diverse physiological processes. Advances in high-throughput non-invasive assays, use of forward- and inverse-genetic approaches, and powerful algorithms are allowing quantitation of variation and detection of genes associated with circadian dynamics. Circadian rhythms and phytohormone pathways in response to endogenous and exogenous cues have been well documented the model plant Arabidopsis. Novel allelic variation associated with circadian rhythms facilitates adaptation and range expansion, and may provide additional opportunity to tailor climate-resilient crops. The circadian phase and period can determine adaptation to environments, while the robustness in the circadian amplitude can enhance resilience to environmental changes. Circadian rhythms in plants are tightly controlled by multiple and interlocked transcriptional–translational feedback loops involving morning (CCA1, LHY), mid-day (PRR9, PRR7, PRR5), and evening (TOC1, ELF3, ELF4, LUX) genes that maintain the plant circadian clock ticking. Significant progress has been made to unravel the functions of circadian rhythms and clock genes that regulate traits, via interaction with phytohormones and trait-responsive genes, in diverse crops. Altered circadian rhythms and clock genes may contribute to hybrid vigor as shown in Arabidopsis, maize, and rice. Modifying circadian rhythms via transgenesis or genome-editing may provide additional opportunities to develop crops with better buffering capacity to environmental stresses. Models that involve clock gene‒phytohormone‒trait interactions can provide novel insights to orchestrate circadian rhythms and modulate clock genes to facilitate breeding of all season crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Genes and networks associated with circadian rhythms
Arabidopsis thaliana, a model plant in research, possesses the most extensively characterized circadian network (Fig. 1A and B). Through forward and reverse molecular genetics, it has been elucidated that the circadian rhythm in plants is tightly controlled by multiple interlocked transcriptional–translational feedback loops (TTFL) (Hernando et al. 2017; McClung 2019; Patnaik et al. 2022). A key TTFL is composed of the genes Circadian Clock Associated 1 (CCA1), Late Elongated Hypocotyl (LHY), and Timing of CAB2 Expression 1 (TOC1). CCA1 and LHY are partially redundant MYB-related Transcription Factors (TFs) that belong to the Reveille (RVE) gene family, where both are expressed at dawn; i.e. during the onset of the light phase of day–night cycle (Lu et al. 2009; Schaffer et al. 1998; Wang et al. 1997). In contrast, TOC1 belongs to the pseudo-response regulator (PRR) gene family expressed in the evening (Gendron et al. 2012). Both CCA1 and LHY proteins can hetero- and homo-dimerize (Lu et al. 2009; Yakir et al. 2009), and negatively regulate gene expression by interacting with the “Evening Elements” (EE, AAAATATCT) and “CCA1-binding sites” (CBS) on the promoter region of its own genes, of TOC1 and other circadian clock genes (Carre and Kay 1995; Harmer et al. 2000; Nagel et al. 2015; Wang et al. 1997). TOC1 also acts as a transcription repressor of itself and other circadian regulatory components (Gendron et al. 2012; Huang et al. 2012; Pokhilko et al. 2012). TOC1 represses the expression of CCA1 by interacting with a Teosinte Branched1-Cycloidea-PCE (TCP) protein, the CCA1 Hiking Expedition (CHE) TF (Pruneda-Paz et al. 2009). Thus, CCA1/LHY and TOC1 represent the core negative TTFL in the A. thaliana circadian clock, where the morning-expressed CCA1/LHY represses the transcription of the evening-expressed TOC1, while TOC1 inhibits the expression of CCA1/LHY during the evening.
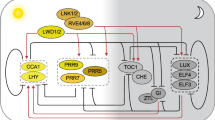
Circadian oscillator components. A Extended model representation of the components of the circadian oscillator. B Simplified model representation of the components of the circadian oscillator. Solid black lines ending with dashes represent the transcriptional repression. C Graphic representation between the hormonal response and the circadian oscillator. The relative timing of action to every component is illustrated in the upper part of the extended model, from left to right during a day–night cycle. Transcriptional repressors are filled in dark blue colour, while transcriptional activators are filled in green colour. Solid lines represent direct regulatory effects while dashed lines represent indirect (or possibly direct but not experimentally determined) effects. Black lines ending with dashes represent the transcriptional repression; green lines ending with arrows represent the transcriptional activation. The circadian repressor components are Circadian Clock Associated 1 (CCA1); Late Elongated Hypocotyl (LHY); Timing of CAB2 Expression 1 (TOC1); Pseudo-Response Regulator 9 (PRR9), PRR7 and PRR5; evening complex (EC) components formed by LUX Arrhythmo (LUX) or Brother of LUX Arrhythmo (BOA), Early Flowering 3 (ELE3), and ELE4; Cold-Regulated 27 (COR27) and COR28; CCA1 Hiking Expedition (CHE). The circadian activation components are Reveille (RVEs); Night Light-Inducible and Clock-Regulated 1 (LNK1), and LNK2; Light-Regulated WD1 (LWD1) and LWD2; Teosinte Branched1-Cycloidea-PCE 20 (TCP20), and TCP22. Proteins involved in brassinosteroids (BRs) circadian regulations are bri1-EMS-Suppressor 1 (BES1) and Topless (TPL). Proteins involved in the Abscisic acid (ABA) circadian regulation are biosynthesis 9-cis-epoxycarotenoid dioxygenase enzymes 3 (NCED3) and MYB96. In the Jasmonic acid (JA) circadian regulation, a basic helix-loop-helix-leucine zipper transcription factor (MYC2) is a key component. Regarding auxin regulation, RVE1 (a member of the RVE transcription factor family but not involved in the core circadian regulation) as well as the auxin biosynthetic gene YUCCA8 (YUC8) are involved
In addition to TOC1, other PRR proteins are involved in circadian rhythm oscillations. PRR9, PRR7, PRR5 and TOC1 are consecutively expressed with moderate differences in their peak expression (Gendron et al. 2012; Liu et al. 2013; Matsushika et al. 2000; Michael et al. 2003; Nakamichi et al. 2010, 2012). PRR9, PRR7 and PRR5 also act as transcription repressors, and together with TOC1, repress the previously expressed PRRs and collectively restrict the transcription of CCA1 and LHY to a period during the dawn (Liu et al. 2013; Nakamichi et al. 2010, 2012). In turn, CCA1/LHY transcriptionally represses PRR9 and PRR7 during the dawn (Adams et al. 2015; Fogelmark and Troein 2014; Greenwood et al. 2022; Joanito et al. 2018; Kamioka et al. 2016; Shalit-Kaneh et al. 2018). Another key component of the clock that is transcriptionally repressed by CCA1/LHY and TOC1 is the evening complex (EC) (Gendron et al. 2012; Huang and Nusinow 2016). The EC is composed of the two plant-specific proteins Early Flowering 3 (ELF3) and Early Flowering 4 (ELF4) and the Lux Arrhythmo (LUX), a MYB-like TF that belongs to the GARP protein family or its close homolog Brother of Lux Arrhythmo (BOA) (Dai et al. 2011; Herrero et al. 2012; Nusinow et al. 2011). The EC complex is expressed after dusk and acts as a transcriptional repressor that binds to the promoter regions of PRR7, PRR9 and LUX (Chow et al. 2012; Dixon et al. 2011; Helfer et al. 2011). LUX (or BOA) is the EC component with DNA-binding capability, since ELF3 and ELF4 do not bind to DNA (Huang and Nusinow 2016). Other corepressors involved in the TTFL are COLD-Regulated 27 (COR27) and COR28, which are repressed by CAA1 and repress PRR5 and TOC1 (Li et al. 2016; Wang et al. 2013; Rawat et al. 2011). These RVEs interact with the coactivators Night Light-Inducible and Clock-Regulated 1 (LNK1) and LNK2 to form a complex and activate the transcription of TOC1, PRR5 and ELF4 (Rugnone et al. 2013; ** system, circadian clock genes generate approximately 24-h rhythms in physiology and behavior. The detection of clock genes can be based on two complementary approaches. The forward-genetic approach is based on arhythmic mutants that produce rhythms deviating from 24-h cycles. Although this approach has been instrumental for revealing the detailed molecular mechanisms of circadian rhythms in model systems and various outcrossing species (King and Takahashi 1996; Miyoshi et al. 2019), it has some significant drawbacks. First, its translational relevance for a broad spectrum of species and biological processes is challenging since only a limited number of mutations can be identified. Also, many clock genes are not evolutionarily conserved; e.g., some clock genes are species-specific (Calixto et al. 2016). This suggests that ideally, detection of clock genes should be based on individual species. To overcome these drawbacks, a forward-genetic approach has been developed which takes advantage of genome-wide map** and association genetics available for a range of species (Jones et al. 2019). Such an approach could identify a more complete set of clock genes. For example, using a map** population, a number of QTL for circadian clock parameters, such as period, phase, and amplitude, were identified in Arabidopsis thaliana (Rubin et al. 2019). A review of progress in clock QTL map** and its application to understanding the genetic architecture of plant adaptation is available (Anwer and Quint 2022).
Most clock map** attempts have been based on genotype–phenotype association analysis using markers. However, the results from these analyses remain an approximation of reality as the circadian rhythm is complex, involving interactions of many genes with environment. A computational algorithm to map a complete set of clock genes using association genetics has been developed (Sun et al. 2021). This algorithm is powerful in terms of its capacity to reconstruct omnigenic interactome networks for rhythmic traits (Sun et al. 2021). Omnigenic theory predicts that all genes that an organism may carry can be involved in controlling a complex trait developed through gene–gene interaction networks (Boyle et al. 2017). Factoring epistasis interactions (i.e., epiGWAS) could resolve some of the missing heritability for complex traits often found in GWAS or biparental populations (Slim et al. 2020). The algorithm can coalesce all genes into bidirectional, signed, and weighted networks, allowing for the characterization of a roadmap of how each and every gene affects circadian rhythms through direct and indirect pathways. It can also discern how each gene functions. First, some clock genes are significant due to their own strong intrinsic capacity, thus implying that these genes govern circadian rhythms without relying on the promotion of other regulators. Second, the significance of some genes is due to favorable regulation involving other genes. The practical use of these two gene types to improve rhythmic traits should be based on different strategies. In addition, some genes are detected to be insignificant not because their effects are inherently small but because they are inhibited by negative regulators. Thus, the effects of these insignificant genes can be released by knocking down the negative regulators. In theory, this is also an approach for retrieving missing heritability, a common phenomenon detected in current genome-wide association studies. The statistical robustness of this approach has been confirmed (Sun et al. 2021) and also its biological value has been validated by analyzing periodic gene expression data of the malaria parasite Plasmodium falciparum (Smith et al. 2020) and growth trajectory data under contrast environmental conditions in a woody plant, Populus euphrates (Wang et al. 2021).
Non-invasive, high throughput assays to measure circadian rhythms
Lack of high throughput protocols has hindered progress in investigating circadian dynamics where such assays were either manually intensive, or of low throughput, technologically expensive, or required genetic modification (Ford et al. 2016; Nagano et al. 2012; Sugiyama et al. 2001; Xu et al. Phytohormones are signaling molecules present in plants that occur in extremely low concentrations and play a major role in plant growth, development, reproduction, and fitness. Abscisic acid (ABA), ethylene (ET), jasmonates (JA), and salicylic acid (SA) are some of the stress-responsive phytohormones, while auxin (AUX), brassinosteroids (BRs), cytokinins (CK), and gibberellins (GA) are growth promoting phytohormones (Gray 2004; Verma et al. 2016). Phytohormones control specific features of the plant–circadian system, though in distinct ways. CK delays circadian phase, AUX regulates circadian amplitude and clock precision, and BR and ABA modulate circadian periodicity. This indicates that plants have multiple input/output feedbacks to adjust the clock to external signals and accurately maintain the clock system (Hanano et al. 2006). The circadian clock interconnects with an intricate network of different pathways, including phytohormones (Singh and Mas 2018). Arabidopsis has been extensively investigated to unlock the molecular basis of connecting the circadian rhythms and phytohormone pathways in response to endogenous and exogenous cues (Fig. 1C). A complex interaction between the circadian clock and ABA pathways in Arabidopsis regulates plant responses to abiotic stress tolerance. Late Elongated Hypocotyl (LHY) transcription factor (TF) modulates ABA biosynthesis and ABA responses, which drives the rhythmic accumulation of ABA to ensure peak accumulation of phytohormones when water deficit is most severe. LHY gene represses 9-cis-epoxycarotenoid dioxygenase enzymes, a rate-limiting step of ABA biosynthesis. Plants overexpressing LHY show reduced levels of ABA under drought, whereas loss-of-function mutants exhibited altered rhythm of ABA accumulation. LHY also binds the promoter of multiple components of ABA signaling pathways and regulates responses downstream of the phytohormone. LHY promotes the expression of ABA-responsive genes involved in stress tolerance while inhibiting the negative effect of ABA on seed germination and plant growth (Adams et al. 2018). Circadian clock gates respond to environmental signals to maximize plant fitness. Clock gating is the clock’s regulation of signaling pathways in a manner that the same stimuli given at different time points result in different responses, usually different intensities, which help plants to reduce stress responses at certain times of the day to ensure optimized growth. The MYB96 TF connects with the clock oscillator to shape the circadian gating of ABA responses by directly binding to the TOC1 promoter to regulate its expression as evidenced in Arabidopsis myb96 and toc1-3 mutants. CCA1 may also regulate MYB96-TOC1 function to alter its circadian expression. Thus, a complex circuitry of CCA1-MYB96-TOC1 regulatory interactions provides the mechanistic basis to connect circadian clock and stress signaling to optimize plant defense and fitness (Lee et al. 2016). BRs control many physiological processes via extensive cross-talks with diverse signaling networks. The BR-activated TF bri1-EMS-Suppressor 1 (BES1) together with TPL represses CCA1 and LHY expression at night by binding to their promoters. This repression by BR treatment is compromised in bes1-ko and tpl8 mutants. Consistent BRs treatment for an extended periods shorten the circadian period, where BR-induced rhythmic shortening was impaired in bes1-ko and tpl8 single mutants as well as in the double mutant, cca1-1lhy-21, providing evidence that BES1/TPL-CCA1/LHY module shapes circadian gating of BR signaling in plants (Lee et al. 2020). Leaf senescence is associated with plant ageing or induced by external stresses to relocate nutrients from aging tissues to juvenile and reproductive organs. JA, an endogenous signal, induces leaf senescence (Sobieszczuk-Nowicka et al. 2018). The evening complex (EC), a core component of the circadian oscillator, negatively regulates leaf senescence. EC is involved in JA signaling and response as evidenced by accelerated leaf senescence in EC mutants upon JA induction. EC binds MYC2 promoter to repress its expression. MYC2 is the key activator of JA-induced leaf senescence as in EC triple (myc2 myc3 myc4) mutants. The accelerated JA-induced leaf senescence in EC mutant is abrogated by myc2 myc3 myc4 triple mutation, providing direct evidence that a core component of the circadian clock gates JA signaling to regulate leaf senescence (Zhang et al. 2018b). JA signaling regulates multiple outputs of plant defense and growth to minimize adverse effects of growth-defense trade-offs in complex environments (Li et al. 2022). Auxin plays a central role in plant growth and development in response to environmental cues. RVE1 is a Myb-like clock regulated TF homologous to CCA1 and LHY. Its inactivation has no effect on circadian rhythmicity but instead causes a growth phenotype, indicating it as a ‘clock output’ that affects plant development. CCA1 regulates growth via bHLH TFs PIF4 and PIF5. However, RVE1 acts independent of PIF4 and PIF5. RVE1 regulates auxin levels by promoting its production during the day but not at night. It positively regulates the expression of auxin biosynthesis gene YUC8 to promote growth. Thus, YUC8 connects two signaling networks to coordinate plant growth with rhythmic changes in the environment (Rawat et al. 2009). OsCCA1, a key clock component in rice, confers tolerance to drought, osmotic, and salinity stresses. There are 692 direct transcriptional target genes of OsCCA1 where, genes involved in ABA signaling are substantially enriched. OsCCA1 binds the promoters of OsPP108 and OsbZIP46 to activate their expression. Oscca1 null mutants display enhanced sensitivity to ABA signaling. Thus, abiotic stress adaptation by OsCCA1 is orchestrated by ABA signaling, linking the circadian clock with ABA signaling in rice (Wei et al. 2022). The OsRCAR10-OsABI5-OsPRR95 feedback loop regulates ABA signaling to fine-tune seed germination and seedling growth, indicating molecular link between ABA signaling and circadian clock in rice (Wang et al. 2022a). The presence of natural variation for circadian rhythms enables plant species to adapt to a wide range of latitudes with variable climates. The examples include variation in circadian period along a latitudinal gradient in annual population of common yellow monkeyflower (Mimulus guttatus) and domesticated soybean crop (Greenham et al. 2017); period length covaries with both geography and population substructure among Swedish Arabidopsis accessions (Rees et al. 2021); free running period (FRP)-dependent phase phenotype underlies photoperiod (short day) adaptation of Japanese duck weed (Lemna aequinoctialis) (Muranaka et al. 2022); and deceleration in circadian clock due to phase delay caused by allelic variation of the tomato homolog of the Arabidopsis EID1 expanded adaptation of domesticated tomatoes in farthest environment in Europe (long day photoperiod) far away from the center of origin (Müller et al. 2016). Many crop wild relatives harbour genetic variation which enables them to flourish in harsh environments. Circadian rhythms in wild barley populations from ecogeographical locations of Southern Fertile Crescent in the Middle East, for example, revealed variation in circadian period length that correlated with temperature and aspects at the sites of plant origin, while the amplitudes of the rhythms correlated with soil composition. These data suggest that variation in environmental parameters exerts selection pressure on circadian rhythms and affects species adaptation (Dakhiya et al. 2017). A comparison of circadian clock rhythmicity and crop performance involving diverse barley lines (wild ancestors, landraces and breeding lines) under optimal and heat-stressed conditions revealed significant loss of thermal plasticity in circadian rhythms during domestication for output genes, while temperature compensation mechanisms was maintained in the core clock (Prusty et al. 2021). Domestication and artificial selection brought substantial changes in the developmental and circadian clock of plants possibly due to restructuring of plant metabolic timekee** (Siqueira et al. 2023). Wild relatives of domesticated crops are an important genetic resource for identifying and exploiting such variations in crop breeding. Rapeseed and wheat cultivars, widely grown, also show variations in circadian periods (Rees et al. 2019). Daily rhythms of gene expression are critical to ensure that biological processes occur at the optimal time of the day (TOD) for traits of agricultural significance. A gene expression study in rice grown in the field environment and temporally monitored for an entire growing season revealed that most genes have a TOD-specific expression over the developmental time course. The thermocycle genes have stronger cue for TOD expression than the photocyclic genes. The core circadian clock genes, except for two grass paralogs of ELF3, display consistent TOD expression over the season, while ELF3 paralogs display distinct phases based on the interaction between thermo- and photo-cycles (Michael 2022). The PRRs gene family in soybean exhibits rhythmic expression under LD and SD. The expression of most of the PRR genes is higher under LD in leaves. An investigating of the effects of natural variation in GmPRR alleles on soybean adaptation involving 207 germplasm accessions from China and USA. The majority of non-synonymous mutations in the coding region are associated with flowering time, while nonsense mutations—because of deletion of CCT domain—relate to early flowering (Wang et al. 2022b). Most of the Northeast China germplasm have haplotypes associated with early flowering, whereas those associated with late flowering are from lower latitudes. Thus, the PRR gene family in soybean provides a circadian clock-controlled flowering pathway and means to breed new germplasm adapted to specific daylength (Wang et al. 2022b). Variation in circadian rhythm plasticity, denoting changes in period and amplitude under distinct temperatures, has been linked to both the maternal organellar genomes (plasmotype) and several nuclear loci in wild barley (Bdolach et al. 2019). Despite these findings, the molecular and biochemical basis of such cytonuclear interactions remain to be elucidated, as well as explored in other crops and models. In short, assessing and exploiting functional allelic diversity in circadian rhythms and clock gene expression on a range of diversity panels of food crop germplasm may provide opportunity to tailor climate-resilient crops. Climate change and variability can affect plant productivity. The increase in temperature can accelerate the reproductive phase in plant life cycle, thus reducing harvests (Parthasarathi et al. 2022). It has been noted that plant development affected by photoperiod uses gene networks that respond to changes in the light/dark cycle length (Osnato et al. 2022). Such circadian clocks, which are important for photoperiod-dependent flowering in high latitudes, are also influenced by temperature changes, thus triggering seasonal responses (Oravec and Greenham 2022). Adapting crops to changing climate in high latitudes, as evidenced in rice and wheat, must consider the interactions between photoperiod, temperature, and water availability. Transcriptome to research has provided insights on increased expression due to high temperature of core clock genes such as CCA1, Gigantea-like protein (GI), PRR59, PRR73, PRR95, and HvLUX (an ortholog of the Arabidopsis circadian gene Lux Arrhythmo, also known as Phytoclock1) in barley (Ford et al. 2016). GI and PRR59 act rapidly when temperature increases, while both CCA1 and PRR73 respond when temperature decreases. The elf3 variant of barley lacks the GI and PRR responses due to temperature changes, suggesting that GI and PRR depend on a functional ELF3 (Ford et al. 2016). Thus, upregulation of core clock genes is pivotal in plants’ adaptation to temperature variations and the maintenance of circadian rhythms. HvLUX1 (under purifying selection) seems to underly the early maturity 10 (eam10) mutant in barley (Campoli et al. 2013). The gene eam10 leads to circadian defects and interacts with the pseudo-response regulator Photoperiod-H1 (Ppd-H1) gene—a homolog of Arabidopsis AtPRR7—to speed flowering under both long and short photoperiods. The emerging knowledge provides new insights for further genetic modification either through crossbreeding or genetic engineering. Modifying photoperiod through sexual hybridization, transgenics or gene editing will enlarge the range of adaptation in some daylength sensitive crops. For breeders, circadian genes and networks have not been a major focus of artificial selection efforts. Despite this, many agronomic traits such as sensitivity to light/dark cycles (Creux and Harmer 2019; Okada et al. 2017), abiotic stress tolerance (Bonnot et al. 2021; Sharma et al. 2022), flowering and maturation time (Creux and Harmer 2019), plant architecture (Rubin et al. 2018), and yield (Steed et al. 2021) are controlled or affected by components of the circadian clock. Thanks to advances in comparative omics, it has been shown that multiple components of the circadian clock are altered in cultivated crops, when compared to their wild relatives (McClung 2021; Steed et al. 2021). This has occurred even though the domestication and breeding processes were performed without having any explicit focus or knowledge about circadian rhythms. Recent research has been focused on understanding and manipulation of circadian clock components as tools for plant breeding and adaptation. For example, in tomato, a 3 bp deletion in the gene Empfindlicher imdunkelroten Licht protein 1 (EID1), which codes for a Phytochrome A-associated F-box protein, in addition to a near-complete deletion of Night Light-Inducible and Clock-Regulated 2 (LNK2; orthologue to AtLNK2) produces a lengthening of the clock period (Müller et al. 2016, 2018). These modifications enabled tomatoes from the equatorial region in South America to adapt from day-neutral circadian rhythms to the longer photoperiod of Europe and Mesoamerica (Müller et al. 2016, 2018). Another example of modifications in the circadian components, that allowed adaptation to high-latitude short-season regions, are mutations in the early maturity (EAM8) gene from barley (Faure et al. 2012) and in the earliness per se (Eps3) from wheat (Gawroński et al. 2014), which are homologs of the Arabidopsis thaliana EC genes, ELF3 and LUX, respectively (Fig. 2). These mutations in barley and wheat not only affect flowering time but also alter the circadian amplitude, period and phase (Faure et al. 2012; Gawroński et al. 2014). In a similar manner, allelic variations in O. sativa circadian heading-date genes (Hd genes not found in Arabidopsis), Grain Number, Plant and Heading Date 7 (Ghd7), Heading Date 1 (Hd1), Early Heading Date 1 (Ehd1) and Early Heading Date 4 (Ehd4) have contributed to adaptation of rice to higher latitudes (Cui et al. 2020; Gao et al. 2013, 2019). This could reveal that not only amplitude is an important factor in the resilience to changes in the environment, but also the plasticity of the rhythm itself. Heterosis or hybrid vigor refers to the superior performance an F1 hybrid over its parents. Polyploidy (whole genome duplication) provides evolutionary advantages for plant growth, development, and adaptation. Some Arabidopsis allotetraploids and hybrids relative to their diploid parents (Arabidopsis thaliana and A. arenosa), can display vigour for morphological traits. Epigenetic modification of the circadian clock genes CCA1, LHY and their reciprocal regulators TOC1 and GI leads to changes in the expression of downstream genes and pathways. Epigenetic repression of CCA1 and LHY during the day induces TOC1, GI, and downstream genes harboring CCA1 binding site (CBS) 13. Allotetraploids and F1 hybrids produce more chlorophyll and starch than parents in the same environment. Single cca1 mutants, double cca1 lhy, and daily repression of cca1 in TOC1:cca1-RNAi transgenic plants, led to increased expression of downstream genes, chlorophyll, and starch content. Notably, constitutive expression of CCA1 or ectopically expressed TOC1:CCA1 had opposite effects. These studies suggest that hybrids and allopolyploids gain advantages via circadian-mediated physiological and metabolic pathways that lead to growth vigor and increased biomass (Ni et al. 2009). Parent-of-origin effects on biomass heterosis are associated with altered CCA1 expression amplitudes, which are associated with methylation levels of CHH (where H = A, T, or C) sites in the promoter region. The direction of rhythmic expression and hybrid vigor is reversed in reciprocal F1 hybrids involving mutants defective in the RNA-directed DNA methylation but not in maintenance methylation pathways. Such parent-of-origin effects on circadian regulation and heterosis is established during early embryogenesis and maintained throughout growth and development (Ng et al. 2014). Plant immunity often causes growth-defense trade-offs. CCA1 confers heterosis for bacterial defense in hybrids without yield penalty, and even enhances the growth heterosis of hybrids under pathogen infection in Arabidopsis. Its genetic perturbation abrogated heterosis for both defense and growth in hybrids. The improved heterosis for defense and growth of hybrids at different time points (dawn, dusk) of the day is achieved when both resistance and growth are the most effective and have the least fitness cost. Plants promote defense heterosis prior to salicylic acid burst by rhythmically enhancing growth vigor during infections in a diurnal manner (Yang et al. 2021). A comparative assessment of transcript profiles of super-hybrid rice ‘LY2186’ and its parents at multiple time points for 2 day/night cycles identified 1552 rhythmic differentially expressed genes, RDGs (Li et al. 2021). The day-phased RDGs were associated with photosynthesis and the night-phased RDGs were involved in stress response, the major contributor to heterosis in ‘LY2186’. The circadian-related RDGs were core components in both phases (day and night) and primarily regulate downstream genes involved in photosynthesis, starch synthesis, plant hormone signal transduction, and other pathways. RDGs (72% of 282 RDGs) mapped onto yield-related QTL could be considered as candidate genes, which after functional validation, may be deployed in hybrid rice breeding (Li et al. 2021). An integrative profiling of metabolome and proteome in maize hybrids and their inbred parents revealed diurnal regulation of several metabolites and proteins. Also, key enzymes in photosynthetic and photorespiratory pathways and metabolites in C assimilation had nonadditive abundance and mild metabolic heterosis in hybrids. Amino acids display negative mid-parent heterosis (MPH), while sugars, alcohols, and nucleotide metabolites had positive MPH. The daily changes in metabolites and proteins, though small, may accelerate hybrid growth. Photosynthetic pathway metabolites have positive MPH, whereas photorespiratory pathway metabolites have negative MPH, which correspond to nonadditive protein abundance and key enzyme activities in the respective pathways (Li et al. 2020). Greater levels of C fixation and starch accumulation are associated with altered temporal gene expression of diurnally upregulated ZmCCA1a in maize. CCA1a homologs, ZmCCA1a and ZmCCA1b, are diurnally upregulated in hybrids. ZmCCA1 expression complements the cca1 mutant phenotype in Arabidopsis. ZmCCA1b overexpression disrupts circadian rhythms and biomass heterosis due to reduced chlorophyll content and plant height. Reduced stem height results from reduced node elongation but not the total number of nodes. The height phenotype is less severe in the field than in greenhouse. Enhanced light and/or metabolic activities in the field compensates for altered circadian regulation, and activation of morning-phased genes in the hybrids promotes photosynthesis and growth vigor (Ko et al. 2016). Transition from vegetative to reproductive phase, photoperiod-induced flowering, plant height, biomass and pod/seed traits, and storage roots in vegetatively propagated crops or tuberization are major agronomic traits influencing productivity of food crops. In addition to trait-responsive genes (i.e., core genes having identifiable direct impact on trait phenotype), several of these traits are also influenced by variation in circadian rhythms and clock genes. Transition from vegetative to reproductive stage is critical to seed yield. Photoperiod pathways regulate flowering in rice, by synchronizing the exogenous environmental and the endogenous signaling cascades of photoreceptors, the circadian clock, and floral integrator genes. A diurnal rhythmic expression of OsLUX with the peak at dusk promoted flowering via the expression of genes associated with the circadian clock and the output integrators of photoperiodic flowering. OsLUX together with OsELF4a and OsELF3a or OsELF3b form two evening complexes (ECs), of which OsLUX-OsELF3a-OsELF4a predominantly promote photoperiodic flowering. OsELF4a alone also promotes flowering. Loss of OsELF4a, unlike OsLUX, marginally influences flowering under short day (SD), but markedly delays flowering under long day (LD) conditions (Cai et al. 2022). Rice yields are predominantly determined by tillering. A regulatory loop involving the circadian clock, sugar, and strigolactone (SL) pathway regulates tiller bud and panicle development. OsCCA1, the circadian clock gene, positively regulates expression of SL pathway genes, OsTB1, D14 and IPA1 to repress tiller-bud outgrowth. OsCCA1 overexpression or downregulation, respectively, reduces or increases tiller numbers, while manipulating OsPRR1 expression causes opposite effects. OsCCA1 also regulates IPA1 expression to mediate panicle and grain development. OsTB1, D14, and IPA1 act downstream of OsCCA1. Sugars affect OsCCA1 negatively in roots and tiller buds but positively in the shoots to promote tiller-bud outgrowth. The circadian clock integrates sugar responses and the SL pathway to regulate tiller and panicle development (Wang et al. 2020a). Optimizing crop duration to the available growing season is a major breeding target to match crop adaptation with the target environment. Variation in circadian-clock associated genes affects flowering in winter wheat cultivars in United Kingdom and European (Wittern et al. 2023). Early flowering 3 (ELF3) is a candidate gene for earliness per se (Eps) at D1 and B1 loci in wheat genome. A SNP within the coding region of TaELF3-B1 is the causal polymorphism at the Eps-B1 locus. Deletion of the Eps-D1 locus encompassing TaELF3-D1 is the basis of an allele that lies within an introgression region containing an inversion relative to the Chinese Spring D genome. Circadian rhythm is severely disrupted in emmer wheat (T. turgidum) cv. Kronos that carries loss-of-function alleles at TtELF3 locus, while loss of LUX in bread wheat, an Arabidopsis orthologue, also severely disrupts circadian rhythms. Thus, both ELF3 and LUX are part of the wheat circadian oscillator, where they integrate the wheat EC (Rees et al. 2022; Steed et al. 2021). Indeed, there exists sufficient allelic diversity within the three wheat ELF3 homoeologous to apply selection; i.e., delay or advance flowering, without adverse pleiotropic alterations to circadian rhythms in wheat (Wittern et al. 2023). The ‘j’ allele in soybean delays flowering and enhances yield of long-juvenile soybean cultivars under SD conditions. ‘Huaxia-3’ (HX3) is a long-juvenile soybean cultivar with a loss-of-function allele j for the J gene. Transcriptome analysis of HX3 relative to the transgenic line overexpressing J gene unfolded 31 and 2311 differentially expressed genes, respectively, in HX3 under SD and LD conditions, with significantly enriched circadian rhythm pathway in HX3 under SD. GmELF3a and FT genes, GmFT2a and GmFT5a, were downregulated, while GmFT4 was upregulated under SD in HX3. FT homolog GmFT4, in contrast, was downregulated while GmFT1a was upregulated under LD in HX3, thereby suggesting that FT homologs may cause delay in flowering of long-juvenile soybean under SD (Sapey et al. 2022). A previous report using a soybean hairy root expression system to monitor endogenous circadian rhythms and the sensitivity of circadian clock to environmental stimuli revealed that a quadruple mutant deficient in GmLCLs orthologs resulted in extreme short-period circadian rhythm and late flowering in soybean. Thus, morning phased GmLCLs act constitutively to maintain rhythmicity, while their absence delays vegetative to reproductive phase transition in soybean (Wang et al. 2020b). Plants with indeterminate inflorescences produce more flower structures than those with determinate type inflorescences. Decoupling floral primordia initiations from their maturation into grains would mean more resources available for the plants to invest in other important physiological functions. Flowering time genes in barley regulate initiation of floral primordia, while floral growth is specified by light signaling, chloroplast, and vascular development orchestrated by CCT MOTIF FAMILY 4 (HvCMF4), which is expressed in the inflorescence vasculature. Mutations in HvCMF4 increase primordia death and pollen failure by reducing rachis greening and limit energy supply to floral tissues. HvCMF4 senses light and connects with vascular-localized circadian clock to coordinate floral initiation and survival. Pyramiding beneficial alleles for both primordia number and survival may increase grain production in barley, caused by erratic stresses due to climate change and variability (Huang et al. 2023). Several clock genes that impact circadian rhythms and abiotic stress adaptation are summarized herewith in diverse crops (Fig. 3). Melatonin (N-acetyl-5-methoxy-tryptamine) is a key hormone that functions as indole-3-acetic acid like hormones in mammals and plants, thereby regulating gene expression affects circadian rhythms, plant growth and development, and stress tolerance (Fan et al. 2018). Abiotic stress impacts the expression pattern of key melatonin biosynthetic genes. In rice, drought stress under light and dark conditions damages circadian clock genes, tryptophan decarboxylase (TDC), tryptamine 5-hydroxylase (T5H), and serotonin N-acetyltransferase (SNAT), but retains the functionality (circadian rhythm) of N-acetylserotonin O-methyltransferase (ASMT) gene in wild-type (WT) plants. Downregulation of ASMT expression in OsGI mutation suggests the involvement of the melatonin biosynthetic pathway in the OsGI-mediated circadian regulation. Thus, there exists a coordination between circadian rhythms and abiotic stress with respect to melatonin biosynthesis in rice (Ahn et al. 2021). However, more focused research on the role of melatonin on plant circadian clock may provide evidence-based mechanistic connection between melatonin and the circadian clock. Circadian component effects under abiotic stress condition. This figure represents the effect of the mutation/silencing or overexpression of the circadian genes in the tolerance to different abiotic stresses. Solid lines represent direct regulatory effects while dashed lines represent indirect (or possibly direct but not experimentally determined) effects. Black lines ending with dashes represent the transcriptional repression of the circadian oscillator components. Green lines ending with arrows represent the transcriptional activation of the circadian oscillator components. Red lines ending with dashes represent transcriptional repression from or to the inputs and outputs in regard to the circadian oscillator. Blue lines ending with arrows represent transcriptional activation from or to the inputs and outputs regarding the circadian oscillator. The circadian repressor components in Arabidopsis are Circadian Clock Associated 1 (CCA1); Late Elongated Hypocotyl (LHY); Timing of CAB2 Expression 1 (TOC1); Pseudo-Response Regulator 9 (PRR9), PRR7 and PRR5; evening complex (EC). The circadian activation components in Arabidopsis are Reveille (RVEs); Night light-iducible and clock-regulated (LNKs); Light-regulated WD (LWDs); Teosinte Branched1-Cycloidea-PCE (TCPs). In Rice, OsCCA1 seems to transcriptionally activate protein phosphatase 2C (OsPP2C) and leucine zipper 46 (OsbZIP46), which have been shown to cope with the negative effects of ROS, drought, and salinity. OsPRR73 inhibit the High-affinity K + transporter 2;1 (OsHKT2;1) which is a negative factor in conferring tolerance to saline conditions. Similarly, the receptor for activated C kinase 1A (OsRACK1A) is circadian regulated and has been shown to have a negative effect increasing the sensitivity to salinity Drought accelerates leaf senescence which reduces canopy size, loss in photosynthesis, and reduced yields, while drought-induced delayed leaf senescence (i.e., stay green) contributes to crops fitness and productivity (Rivero et al. 2007). Variation in internal signals and environmental factors contribute to initiation and progression of leaf senescence (Guo et al. 2021; Zhang et al. 2021). ZmVQ52 is mainly expressed in maize leaves and is involved in the circadian pathway genes and photosynthetic pathways. WRKY family proteins (ZmWRKY20, ZmWRKY36, ZmWRKY50, ZmWRKY71) interact with ZmVQ52. ZmVQ52 overexpression accelerated pre-mature leaf senescence in Arabidopsis. Several leaf senescence genes in ZmVQ52 overexpression lines were upregulated by premature leaf senescence in Arabidopsis (Yu et al. 2019). A large number of heat stress-responsive genes exhibit diurnal oscillation in Arabidopsis. Wild-type and mutants of the circadian clock genes (CCA1, LHY, PRR7, PRR9) exposed to heat (37 °C) and moderate cold (10 °C) during Zeitgeber (a rhythmically occurring natural phenomenon which acts as a cue in the regulation of the plant circadian rhythms) time ZT1 (early morning) and ZT6 (afternoon) revealed thousands of genes that were differentially expressed in response to temperature, the time of the day, or the clock mutation. In the morning, ~ 30% more genes were differentially expressed than in the afternoon. Heat stress significantly perturbed the transcriptome. Seventy percent of ~ 3000 DEGs showed time of the day (ZT1 or ZT6) occurrence of the transcriptional response. Changes in the magnitude of transcriptional response were detected in ~ 1400 DEGs shared between ZT1 and ZT6. About 2% of all DEGs had differential responses to temperature stress in the clock mutants. The clock genes CCA1 and LHY seem to have a more profound role than PRR7 and PRR9 in modulating heat stress responses during the day. Thus, time of the day plays a significant role in quantifying heat stress-responsive transcriptomes (Blair et al. 2019). RACK1 is a WD40 type protein involved in multiple signaling pathways, conserved from prokaryotes to eukaryotes, and is involved in diverse biological functions and stress tolerance in plants (Adams et al. 2011; Zheng et al. 2017). OsRACK1A is a circadian rhythm gene involved in regulating salt tolerance in rice. OsRACK1A-suppressed transgenic rice compared to WT maintained low Na+ and high K+ concentrations, accumulate significantly more ABA and its transcripts and stress-inducible genes. Many stress-related genes, including AP2/ERF, were upregulated in OsRACK1A-suppressed transgenic rice (Zhang et al. 2018a). Another clock component, OsPRR73, positively regulates salt tolerance in rice by recruiting HDAC10 which transcriptionally repressed OsHKT2; 1, thus reducing cellular Na+ accumulation. osprr73 null mutants accumulated significantly higher levels of reactive oxygen species and sodium ions, which resulted in reduction in grain size and yield under salt stress (Wei et al. 2021). Overall, circadian rhythmicity and clock genes that respond to abiotic stress, in turn interact with stress response genes to confer tolerance, providing broadened opportunities to develop more sustainable and productive cereal crops. In addition, several candidate clock genes with differential expression levels were identified as potential target for functional validation, for example, StGI, StPRR, and StEFM affecting tuberization in potato (Niu et al. 2022); COR expression in day and night impacting cold tolerance (Lu et al. 2021) and variable expression levels of morning (OsPRRs, OsLHY, OsZTL1) and evening (OsTOC1, OsGI, OsELF3) circadian clock genes in response to drought stress in rice (Li et al. 2017); HvELF3 expression impacting phase and shape of the clock and stress response (Habte et al. 2014) and variable expression of core clock genes (CCA1, GI, PRR59, PRR73, PRR95, and LUX) at higher temperatures (Ford et al. 2016) in barley; or circadian clock genes impacting significantly phase and amplitude in root and shoot under varying water regimes in the monocot model plant Brachypodium distachyon (Gombos et al. 2023). Modifying the circadian clock should be a breeding target to develop cultivars with higher buffering capacity to environmental stresses. Transgenic rice (japonica variety Taipei309) overexpressing CCA1 under the control of TOC1 promoter and containing RNAi construct A that follows circadian rhythm in progeny (T1, T2) plants has a detrimental effect on phenology and seed yield related traits over WT. CCA1 repression under the control of TOC1 promoter and containing RNAi constructs B and C that follows circadian rhythm in progeny plants significantly improve phenology and seed yield related traits over WT. Furthermore, RNAi construct C containing CCA1 derived from the 3’ terminal region of CCA1 generates plants with better phenology than construct B obtained from 5’ terminal region of CCA1. The progeny plants did not differ in chlorophyll content but had distinct rhythmicity, relative to CCA1 expression. Thus, CCA1 under the control of TOC1 promoter offers promise for development of crop cultivars better adapted to variable climates (Chaudhary et al. 2019). CRISPR-Cas9 system has shown promise for plant breeding (Haque et al. 2018). Protein–protein interaction networks of differentially expressed proteins offer opportunities for better understanding of the organism response at the whole proteome level (Zou et al. 2018). A comparative proteomic analysis between CRISPR-Cas9 generated OsPYL9 loss-of-function mutants and WT at the whole proteome level revealed that CRISPR mutants had enhanced drought tolerance and improved grain yield under optimal and water-deficit conditions. Multiple proteins were differentially expressed (184 upregulated and 140 downregulated), with most of the upregulated DEPs related to circadian clock rhythm, drought response, and antioxidant activities in the mutants. DEPs were involved in response to abiotic stimulus and abscisic acid-activated signaling pathways. GIGANTEA, Adagio-like, and pseudo-response regulator proteins revealed higher interaction in protein–protein interaction (PPI) network. Both DEPs and higher PPI network in OsPYL9 mutant affect circadian rhythmicity, and a potential genetic resource and biomarker to breed for increased drought tolerance and improved yield in rice (Usman et al. 2020). Overall, the results summarized in this section indicate the potential of genetic engineering for modifying circadian rhythmicity to enhance stress tolerance and productivity in food crops. The circadian clock that is based on interlocked negative loops of transcriptional activators and repressors is crucial for adjusting crop growth. Genes that maintain the plant circadian clock “ticking” (e.g. morning genes: CCA1, LHY; mid-day genes: PRR9, PRR7, PRR5; evening genes: TOC1, ELF3, ELF4, LUX) are supported by evidence in model plant Arabidopsis and other crops. Clock genes impacting biological rhythms and associated with plant growth, development, reproduction, and stress tolerance, have been identified in staple crops. Likewise, the role of phytohormones in mediating clock and stress-responsive gene expression is known. Maintaining the intricate interactions between circadian rhythms and clock genes‒phytohormones‒trait/stress-responsive genes may open new paths to tailor crops adapted to external environmental cues. Crop adaptation by changing flowering time is a good example of the potential of clock genes, where loss of some segments of circadian clock genes facilitated adaptation of tomatoes to distant growing habitats far from its center of origin (Müller et al. 2016, 2018). The knowledge emerging on circadian clock molecular functioning can facilitate breeding of all seasons’ crops (Fig. 4). However, pursuit of such an approach poses a significant breeding challenge due to various factors; namely, (i) harnessing circadian oscillator genes that confer variation in biological rhythms in crop genepools, (ii) significant bottlenecks to establishing cost-effective high-throughput phenomics to assess variation in functioning of circadian oscillators in early breeding generations, (iii) possible negative trade-offs involving clock oscillator genes, phytohormones, abiotic stresses, and gene expression (i.e., trait- and stress-responsive genes), and (iv) identifying common genetic markers (e.g., SNPs, InDels and structural variation) linked with clock oscillators, phytohormones, and gene expression. Thus, assessing and exploiting functional allelic diversity in core oscillators as well as in trait-and stress-responsive genes can accelerate the development of climate-resilient productive and value-added germplasm for use in crop breeding. The circadian oscillator in plants: integrating environmental and physiological cues to improve adaptation and resilience. The intricate circadian oscillator in plants continually integrates environmental and physiological cues, including signals from phytohormones, natural environments, and abiotic stress conditions. The oscillator responds to these inputs with coordinated output signals, allowing the plant to adjust its physiology and behaviour accordingly. Recent research has revealed that natural allelic variations or induced mutations in genes involved in the regulation of the circadian oscillator can significantly impact how plants perceive inputs or regulate outputs, ultimately leading to improved adaptation to new environments, enhanced tolerance to abiotic stress, and increased crop yields. This figure highlights the importance of understanding the genetic and molecular mechanisms underlying the circadian oscillator’s regulation and how they can be manipulated to improve plant performance and resilienceCircadian control of phytohormone biosynthesis and signaling sha** plants to external environments
Modifying circadian rhythms to develop climate-resilient productive crop cultivars
Novel variation in circadian clock systems
Photoperiod (or temperature) × circadian clock × early flowering gene interaction to enhance crops range adaptation
Severely compromised clock gene expression and clock outputs can favor crop adaptation
Altered circadian rhythms and clock genes contribute to heterosis
Biomass and defense heterosis in Arabidopsis
Yield heterosis in grain crops
Circadian rhythms and clock genes mediating agronomic traits and abiotic stress adaptation
Agronomic traits
Abiotic stress adaptation
Transgenic modification or genome editing to alter circadian rhythms to enhance abiotic stress adaptation and crops productivity
Managing circadian clock for breeding all seasons’ crops
Data availability
No new data were generated for preparation of this review paper. Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Adams DR, Ron D, Kiely PA (2011) RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal 9:22. https://doi.org/10.1186/1478-811X-9-22
Adams S, Manfield I, Stockley P, Carré IA (2015) Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS ONE 10:e0143943. https://doi.org/10.1371/journal.pone.0143943
Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S, Carré IA (2018) Circadian control of abscisic acid biosynthesis and signaling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol 220:893–907. https://doi.org/10.1111/nph.15415
Ahn H-Y, Kim Y-J, Lim Y-J, Duan S, Eom S-H, Jung K-H (2021) Key genes in the melatonin biosynthesis pathway with circadian rhythm are associated with various abiotic stresses. Plants (basel) 10:129. https://doi.org/10.3390/plants10010129
Anwer MU, Quint M (2022) How to detect QTLs in the plant circadian clock. Methods Mol Biol 2398:89–97. https://doi.org/10.1007/978-1-0716-1912-4_8
Bdolach E, Prusty MR, Faigenboim-Doron A, Filichkin T, Helgerson L, Schmid KJ, Greiner S, Fridman E (2019) Thermal plasticity of the circadian clock is under nuclear and cytoplasmic control in wild barley. Plant Cell Environ 42:3105–3120. https://doi.org/10.1111/pce.13606
Blair EJ, Bonnot T, Hummel M, Hay E, Marzolino JM, Quijada IA, Nagel DH (2019) Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Scientific Rep 9:4814. https://doi.org/10.1038/s41598-019-41234-w
Blanca J, Sanchez-Matarredona D, Ziarsolo P, Montero-Pau J, van der Knaap E, Díez MJ, Cañizares J (2022) Haplotype analyses reveal novel insights into tomato history and domestication driven by long-distance migrations and latitudinal adaptations. Hort Res 9:uhac030. https://doi.org/10.1093/hr/uhac030
Bonnot T, Blair EJ, Cordingley SJ, Nagel DH (2021) Circadian coordination of cellular processes and abiotic stress responses. Curr Opin Plant Biol 64:102133. https://doi.org/10.1016/j.pbi.2021.102133
Boyle EA, Li YI, Pritchard JK (2017) An expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–1186. https://doi.org/10.1016/j.cell.2017.05.038
Cai Z, Zhang Y, Tang W, Chen X, Lin C, Liu Y, Ye Y, Wu W, Duan Y (2022) Lux Arrhythmo interacts with ELF3a and ELF4a to coordinate vegetative growth and photoperiodic flowering in rice. Front Plant Sci 13:853042. https://doi.org/10.3389/fpls.2022.853042
Calixto CPG, Simpson CG, Waugh R, Brown JW (2016) Alternative splicing of barley clock genes in response to low temperature. PLoS ONE 11:e0168028. https://doi.org/10.1371/journal.pone.0168028
Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M (2013) HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199:1045–1059
Carre IA, Kay SA (1995) Multiple DNA-protein complexes at a circadian-regulated promoter element. Plant Cell 7:2039–2051. https://doi.org/10.1105/tpc.7.12.2039
Chaudhary S, Kohkhar W, Jabre I, Reddy ASN, Byrne LJ, Wilson CM, Syed NH (2019) Alternative splicing and protein diversity: plants versus animals. Front Plant Sci 10:708. https://doi.org/10.3389/fpls.2019.00708
Chow BY, Helfer A, Nusinow DA, Kay SA (2012) ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signal Behav 7:170–173. https://doi.org/10.4161/psb.18766
Creux N, Harmer S (2019) Circadian rhythms in plants. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a034611
Cui Y, Wang J, Feng L, Liu S, Li J, Qiao W, Song Y, Zhang Z, Cheng Y, Zhang L et al (2020) A combination of long-day suppressor genes contributes to the northward expansion of rice. Front Plant Sci 11:864. https://doi.org/10.3389/fpls.2020.00864
Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, Ruff TG, Beachy RN (2011) Brother of Lux Arrhythmo is a component of the Arabidopsis circadian clock. Plant Cell 23:961–972. https://doi.org/10.1105/tpc.111.084293
Dakhiya Y, Hussien D, Frodman E, Kiflawi M, Green R (2017) Correlation between circadian rhythms and growth in challenging environments. Plant Physiol 173:1724–1734
Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through Early Flowering 3 in Arabidopsis. Curr Biol 21:120–125. https://doi.org/10.1016/j.cub.2010.12.013
Fan J, **e Y, Zhang Z, Chen L (2018) Melatonin: a multifunctional factor in plants. Int J Mol Sci 19:1528. https://doi.org/10.3390/ijms19051528
Farinas B, Mas P (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66:318–329. https://doi.org/10.1111/j.1365-313X.2011.04484.x
Faure S, Turner AS, Gruszka D, Christodoulou V, Davis S, von Korff M, Laurie DA (2012) Mutation at the circadian clock gene Early Maturity 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109:8328–8333. https://doi.org/10.1073/pnas.1120496109
Fogelmark K, Troein C (2014) Rethinking transcriptional activation in the Arabidopsis circadian clock. PLOS Comput Biol 10:e1003705. https://doi.org/10.1371/journal.pcbi.1003705
Ford B, Deng W, Clausen J, Oliver S, Boden S, Hemming M, Trevaski B (2016) Barley (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an Early Flowering 3-dependent manner. J Exp Bot 67:5517–5528. https://doi.org/10.1093/jxb/erw317
Fornara F, de Montaigu A, Sánchez-Villarreal A, Takahashi Y, Loren V, van Themaat E, Huettel B, Davis SJ, Coupland G (2015) The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81:695–706. https://doi.org/10.1111/tpj.12759
Gao H, Zheng X-M, Fei G, Chen J, ** M, Ren Y, Wu W, Zhou K, Sheng P, Zhou F et al (2013) Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet 9:e1003281. https://doi.org/10.1371/journal.pgen.1003281
Gao H, ** M, Zheng X-M, Chen J, Yuan D, **n Y, Wang M, Huang D, Zhang Z, Zhou K et al (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci 111:16337–16342. https://doi.org/10.1073/pnas.1418204111
Gawroński P, Ariyadasa R, Himmelbach A, Poursarebani N, Kilian B, Stein N, Steuernagel B, Hensel G, Kumlehn J, Sehgal SK et al (2014) A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics 196:1253–1261. https://doi.org/10.1534/genetics.113.158444
Geldhof B, Pattyn J, Eyland D, Carpentier S, Van de Poel B (2021) A digital sensor to measure real-time leaf movements and detect abiotic stress in plants. Plant Physiol 187:1131–1148. https://doi.org/10.1093/plphys/kiab407
Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109:3167–3172. https://doi.org/10.1073/pnas.1200355109
Gil K-E, Park C-M (2019) Thermal adaptation and plasticity of the plant circadian clock. New Phytol 221:1215–1229. https://doi.org/10.1111/nph.15518
Gombos M, Hapek N, Kozma-Bognár L, Grezal G, Zombori Z, Gyōrgyey J (2023) Limited water stress modulates expression of circadian clock genes in Brachypodium distachyon roots. Sci Rep 13:1241. https://doi.org/10.1038/s41598-022-27287-4
Gould PD, Diaz P, Hogben C, Kusakina J, Salem R, Hartwell J, Hall A (2009) Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J 58:893–901
Gray WM (2004) Hormonal regulation of plant growth and development. PLoS Biol 2:e311. https://doi.org/10.1371/journal.pbio.0020311
Greenham K, Lou P, Remsen SE, Farid H, McClung CR (2015) TRiP: tracking rhythms in plants, an automated leaf movement analysis program for circadian period estimation. Plant Methods 11:33. https://doi.org/10.1186/s13007-015-0075-5
Greenham K, Lou P, Puzey JR, Kumar G, Arnevik C, Farid H, Wills JH, McClung CR (2017) Genetic variation of plant circadian clock function in natural and agricultural settings. J Biol Rhythms 32:26–34. https://doi.org/10.1177/0748730416679307
Greenwood M, Tokuda IT, Locke JCW (2022) A spatial model of the plant circadian clock reveals design principles for coordinated timing. Mol Syst Biol 18:e10140. https://doi.org/10.15252/msb.202010140
Grundy J, StokerC CIA (2015) Circadian regulation of abiotic stress tolerance in plants. Front Plant Sci 6:648. https://doi.org/10.3389/fpls.2015.00648
Guo Y, Ren G, Zhang K, Li Z, Miao Y, Guo H (2021) Leaf senescence: progression, regulation, and application. Mol Hortic 1:5. https://doi.org/10.1186/s43897-021-00006-9
Habte E, Müller LM, Shtava M, Davis SJ, von Korff M (2014) Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant Cell Environ 37:1321–1327. https://doi.org/10.1111/pce.12242
Hanano S, Domagalska MA, Nagy F, Davis SJ (2006) Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11:1381–1392. https://doi.org/10.1111/j.1365-2443.2006.01026.x
Haque E, Taniguchi H, Hassan MM, Bhowmik P, Karim MR, Śmiech M, Zhao K, Rahman M, Islam T (2018) Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front Plant Sci 9:617. https://doi.org/10.3389/fpls.2018.00617
Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113. https://doi.org/10.1126/science.290.5499.2110
Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) Lux Arrhythmo encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21:126–133. https://doi.org/10.1016/j.cub.2010.12.021
Hernando CE, Romanowski A, Yanovsky MJ (2017) Transcriptional and post-transcriptional control of the plant circadian gene regulatory network. Biochim Biophys Acta Gene Regul Mech 1860:84–94. https://doi.org/10.1016/j.bbagrm.2016.07.001
Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R et al (2012) Early Flowering4 Recruitment of Early Flowering3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24:428–443. https://doi.org/10.1105/tpc.111.093807
Hotta CT (2021) From crops to shops: how agriculture can use circadian clocks. J Exp Bot 72:7668–7679. https://doi.org/10.1093/jxb/erab371
Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekee** is controlled by a cycling activator in Arabidopsis. Elife 2:e00473. https://doi.org/10.7554/eLife.00473
Huang H, Nusinow DA (2016) Into the evening: complex interactions in the Arabidopsis circadian clock. Trends Genet 32:674–686. https://doi.org/10.1016/j.tig.2016.08.002
Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Map** the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336:75–79. https://doi.org/10.1126/science.1219075
Huang Y, Kamal R, Shanmugaraj N, Rutten N, Thirulogachandar V, Zhao S, Hoffie I, Hensel G, Rajaraman J, Moya YAT et al (2023) A molecular framework for grain number determination in barley. Sci Adv 9:eadd0324. https://doi.org/10.1126/sciadv.add03
** S, Su Y, Zhang Y, Song S, Li Q, Liu Z, Ma Q, Ge Y, Liu L, Ding Y et al (2021) Exploring seasonal and circadian rhythms in structural traits of field maize from LiDAR time series. Plant Phenom. https://doi.org/10.34133/2021/9895241
Joanito I, Chu JW, Wu S-H, Hsu C-P (2018) An incoherent feed-forward loop switches the Arabidopsis clock rapidly between two hysteretic states. Sci Rep 8:13944. https://doi.org/10.1038/s41598-018-32030-z
Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, Jeffries AR, Dashti HS, Hillsdon M, Ruth KS et al (2019) Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun 10:343. https://doi.org/10.1038/s41467-018-08259-7
Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by Circadian Clock-Associated1 in the Arabidopsis circadian clock. Plant Cell 28:696–711. https://doi.org/10.1105/tpc.15.00737
Kim W-Y, Ali Z, Park HJ, Park SJ, Cha J-Y, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z et al (2013) Release of SOS2 kinase from sequestration with Gigantea determines salt tolerance in Arabidopsis. Nat Commun 4:1352. https://doi.org/10.1038/ncomms2357
Kim JA, Kim HS, Choi SH, Jang J-Y, Jeong M-J, Lee SI (2017) The importance of the circadian clock in regulating plant metabolism. Int J Mol Sci 18:2680. https://doi.org/10.3390/ijms1812268
King DP, Takahashi JS (1996) Forward genetic approaches to circadian clocks in mice. Cold Spring Harb Symp Quant Biol 61:295–302
Ko DK, Rohozinski D, Song Q, Taylor SH, Juenger TE, Harmon FG, Chen ZJ (2016) Temporal shift of circadian-mediated gene expression and carbon fixation contributes to biomass heterosis in maize hybrids. PLoS Genet 12:e1006197. https://doi.org/10.1371/journal.pgen.1006197
Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP (2012) Circadian Clock-Associated 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA 109:17129–17134. https://doi.org/10.1073/pnas.1209148109
Lee HG, Mas P, Seo PJ (2016) MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci Rep 6:17754. https://doi.org/10.1038/srep17754
Lee GH, Won JH, Choi Y-R, Lee K, Seo PJ (2020) Brassinosteroids regulate circadian oscillation via the BES1/TPL-CCA1/LHY module in Arabidopsis thaliana. iScience 23:101528. https://doi.org/10.1016/j.isci.2020.101528
Legnaioli T, Cuevas J, Mas P (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28:3745–3757. https://doi.org/10.1038/emboj.2009.297
Li X, Ma D, Lu SX, Hu X, Huang R, Liang T, Xu T, Tobin EM, Liu H (2016) Blue Llght- and low temperature-regulated COR27 and COR28 play roles in the Arabidopsis circadian clock. Plant Cell 28:2755–2769. https://doi.org/10.1105/tpc.16.00354
Li J, Liu Y-H, Zhang Y, Chen C, Yu X, Yu S-W (2017) Drought stress modulates diurnal oscillations of circadian clock and drought-responsive genes in Oryza sativa L. Hereditas 39:837–846
Li Z, Zhu A, Song Q, Chen HY, Harmon FG, Chen ZJ (2020) Temporal regulation of the metabolome and proteome in photosynthetic and photorespiratory pathways contributes to maize heterosis. Plant Cell 32:3706–3722. https://doi.org/10.1105/tpc.20.00320
Li Y, Liu Y, Zhou Y, Wei X, Peng Y, Dai Y, Zhang L, Zhu Z (2021) Diurnal transcriptomics analysis reveals the regulatory role of the circadian rhythm in super-hybrid rice LY2186. Genomics 113:1281–1290
Li C, Xu M, Cai X, Han Z, Si J, Chen D (2022) Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int J Mol Sci 23:3945. https://doi.org/10.3390/ijms23073945
Lin X, Fang C, Liu B, Kong F (2021) Natural variation and artificial selection of photoperiodic flowering genes and their applications in crop adaptation. aBIOTECH 2:156–169. https://doi.org/10.1007/s42994-021-00039-0
Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM (2013) Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. The Plant J 76:101–114. https://doi.org/10.1111/tpj.12276
Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM (2009) Circadian Clock Associated1 and Late Elongated Hypocotyl function synergistically in the circadian clock of Arabidopsis. Plant Physiol 150:834–843. https://doi.org/10.1104/pp.108.133272
Lu X, Song S, **ao Y, Fan F, Yan Z, Jia G, Tang W, Peng J (2021) Circadian clock-coordinated response to chilling stress in rice. Env Exp Bot 185:104398. https://doi.org/10.1016/j.envexpbot.2021.104398
Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of Pseudo-Response Regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41:1002–1012. https://doi.org/10.1093/pcp/pcd043
McClung C (2019) The plant circadian oscillator. Biology 8:14. https://doi.org/10.3390/biology8010014
McClung CR (2021) Circadian clock components offer targets for crop domestication and improvement. Genes 12:374. https://doi.org/10.3390/genes12030374
Michael TP (2022) Time of the day analysis over a field grown developmental time course in rice. Plants 12:166. https://doi.org/10.3390/plants12010166
Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302:1049–1053. https://doi.org/10.1126/science.1082971
Miyoshi C, Kim SJ, Ezaki T, Ikkyu A, Hotta-Hirashima N, Kanno S, Kakizaki M, Yamada M, Wakana S, Yanagisawa M et al (2019) Methodology and theoretical basis of forward genetic screening for sleep/wakefulness in mice. Proc Natl Acad Sci USA 116:16062–16067. https://doi.org/10.1073/pnas.1906774116
Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72:353–363. https://doi.org/10.1101/sqb.2007.72.006
Müller N, Wijnen C, Srinivasan A, Ryngajllo M, Oftner I, Lin T, Ranjan A, West D, Maloog JN, Sihna NR, Huang S, Zamir D, Jiménez-Gómez J (2016) Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat Genet 48:89–93. https://doi.org/10.1038/ng.3447
Müller NA, Zhang L, Koornneef M, Jiménez-Gómez J (2018) Mutations in EID1 and LNK2 caused light-conditional clock deceleration during tomato domestication. Proc Natl Acad Sci USA 115:7135–7140. https://doi.org/10.1073/pnas.1801862115
Muranaka T, Ito S, Kudoh H, Oyama T (2022) Circadian-period variation underlies the local adaptation of photoperiodism in the short-day plant Lemna aequinoctialis. iScience 25:104634. https://doi.org/10.1016/j.isci.2022.104634
Nagano AJ, Sato Y, Mihara M, Antonio BA, Motoyama R, Itoh H, Nagamura Y, Izawa T (2012) Deciphering and prediction of transcriptome dynamics under fluctuating field condition. Cell 151:1359–1369. https://doi.org/10.1016/j.cell.2012.10.048
Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112:E4802–E4810. https://doi.org/10.1073/pnas.1513609112
Nakamichi N (2015) Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol 56:594–604. https://doi.org/10.1093/pcp/pcu181
Nakamichi N (2020) The transcriptional network in the Arabidopsis circadian clock system. Genes (basel) 11:1284. https://doi.org/10.3390/genes11111284
Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009) Transcript profiling of an Arabidopsis Pseudo Response Regulator arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50:447–462. https://doi.org/10.1093/pcp/pcp004
Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H (2010) Pseudo-Response Regulators 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22:594–605. https://doi.org/10.1105/tpc.109.072892
Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109:17123–17128. https://doi.org/10.1073/pnas.1205156109
Ng DWK, Miller M, Yu HH, Huang T-Y, Kim E-D, Lu J, **e Q, McClung CR, Chen ZJ (2014) A role for CHH methylation in the parent-of-origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell 26:2430–2440. https://doi.org/10.1105/tpc.113.115980
Ni Z, Kim E-D, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ (2009) Altered circadian rhythms regulate growth vigor in hybrids and allopolyploids. Nature 457:327–331. https://doi.org/10.1038/nature07523
Niu Y, Li G, Jian Y, Duan S, Liu J, Xu J, ** L (2022) Genes related to circadian rhythm are involved in regulating tuberization time in potato. Horticultural Plant J 8:369–380. https://doi.org/10.1016/j.hpj.2021.09.003
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402. https://doi.org/10.1038/nature10182
Okada M, Muranaka T, Ito S, Oyama T (2017) Synchrony of plant cellular circadian clocks with heterogeneous properties under light/dark cycles. Sci Rep 7:317. https://doi.org/10.1038/s41598-017-00454-8
Oravec MW, Greenham K (2022) The adaptive nature of the plant circadian clock in natural environments. Plant Physiol 190:968–980. https://doi.org/10.1093/plphys/kiac337
Osnato M, Cota I, Nebhnani P, Cereijo U, Pelaz S (2022) Photoperiod control of plant growth: flowering time genes beyond flowering. Front Plant Sci 12:805635. https://doi.org/10.3389/fpls.2021.805635
Parthasarathi T, Firdous S, David EM, Lesharadevi K, Djanaguiraman M (2022) Effects of high temperature on crops. In: Kimatu JN (ed) Advances in plant defense mechanisms, vol 18. IntechOpen. https://doi.org/10.5772/intechopen.105945
Patnaik A, Alavilli H, Rath J, Panigrahi KCS, Panigrahy M (2022) Variations in circadian clock organization and function: a journey from ancient to recent. Planta 256:91. https://doi.org/10.1007/s00425-022-04002-1
Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8:574. https://doi.org/10.1038/msb.2012.6
Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323:1481–1485. https://doi.org/10.1126/science.1167206
Prusty MR, Bdolach E, Yamamoto E, Tiwari LD, Silberman R, Doron-Faigenbaum A, Neyhart JL, Bonfils D, Kashkush K, Pillen K et al (2021) Genetic loci mediating circadian clock output plasticity and crop productivity under barley domestication. New Phytol 230:1787–1801
Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, Zhao Y, Ljung K, Harmer S (2009) Reveille1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA 106:16883–16888
Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) Reveille8 and Pseudo-Reponse Regulator5 form a negative feedback loop within the Arabidopsis circadian clock. PLOS Genet 7:e1001350. https://doi.org/10.1371/journal.pgen.1001350
Rees H, Duncan S, Gould P, Wells R, Greenwood M, Brabbs T, Hall A (2019) A high-throughput delayed fluorescence method reveals underlying differences in the control of circadian rhythms in Triticum aestivum and Brassica napus. Plant Methods 15:51. https://doi.org/10.1186/s13007-019-0436-6
Rees H, Joynson R, Brown JKM, Hall A (2021) Naturally occurring circadian rhythm variation associated with clock gene loci in Swedish Arabidopsis accessions. Plant Cell Environ 44:807–820. https://doi.org/10.1111/pce.13941
Rees H, Rusholme-Pilcher R, Bailey P, Colmer J, White B, Reynolds C, Ward SJ, Coombes B, Graham CA, de Barros Dantas LL et al (2022) Circadian regulation of the transcriptome in a complex polyploid crop. PLoS Biol 20:e3001802. https://doi.org/10.1371/journal.pbio.3001802
Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104:19631–19636. https://doi.org/10.1073/pnas.0709453104
Rubin MJ, Brock MT, Baker RL, Wilcox S, Anderson K, Davis SJ, Weinig C (2018) Circadian rhythms are associated with shoot architecture in natural settings. New Phytol 219:246–258. https://doi.org/10.1111/nph.15162
Rubin MJ, Brock MT, Davis SJ, Weinig C (2019) QTL underlying circadian clock parameters under seasonally variable field settings in Arabidopsis thaliana. G3 (bethesda) 9:1131–1139. https://doi.org/10.1534/g3.118.200770
Rugnone ML, Faigón Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Más P et al (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110:12120–12125. https://doi.org/10.1073/pnas.1302170110
Saini R, Jaskolski M, Davis SJ (2019) Circadian oscillator proteins across the kingdoms of life: structural aspects. BMC Biol 17:13. https://doi.org/10.1186/s12915-018-0623-3
Sanchez SE, Kay SA (2016) The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harb Perspect Biol 8:a027748. https://doi.org/10.1101/cshperspect.a027748
Sapey E, Jiang B, Liu L, Yuan S, Wu T, Ibrahim SE, Sun S, Yue Y, Han T (2022) Transcriptome profile of a long-juvenile soybean genotype Huaxia-3 under short and long photoperiod. Plant Mol Biol Rep. https://doi.org/10.1007/s11105-021-01332-4
Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93:1219–1229. https://doi.org/10.1016/s0092-8674(00)81465-8
Shalit-Kaneh A, Kumimoto RW, Filkov V, Harmer SL (2018) Multiple feedback loops of the Arabidopsis circadian clock provide rhythmic robustness across environmental conditions. Proc Natl Acad Sci USA 115:7147–7152. https://doi.org/10.1073/pnas.1805524115
Sharma M, Irfan M, Kumar A, Kumar P, Datta A (2022) Recent insights into plant circadian clock response against abiotic stress. J Plant Growth Regul 41:3530–3543. https://doi.org/10.1007/s00344-021-10531-y
Singh M, Mas P (2018) A functional connection between the circadian clock and hormonal timing in Arabidopsis. Genes (basel) 9:567. https://doi.org/10.3390/genes9120567
Siqueira JA, Batista-Silva W, Zsögön A, Fernie AR, Araújo WL, Nunes-Nesi A (2023) Plant domestication: setting biological clocks. Trends Plant Sci. https://doi.org/10.1016/j.plants.2023.01.009
Slim L, Chatelain C, Azencott C-A, Vert J-P (2020) Novel methods for epistasis detection in genome-wide association studies. PLoS ONE 15:e0242927. https://doi.org/10.1371/journal.pone.0242927
Smith LM, Motta F, Chopra G, Moch JK, Nerem RR, Cummins B, Roche KE, Kellihar CM, Leman AR, Harer J et al (2020) An intrinsic oscillator drives the blood stage cycle of the malaria parasite, Plasmodium falciparum. Science 368:754–759. https://doi.org/10.1126/science.aba4357
Sobiesczuk-Nowicka E, Wrzesiński T, Bagniewska-Zadworna A, Kubala S, Rucińska-Sobkowiak R, Polcyn W, Misztal L, Mattoo AK (2018) Physio-genetic dissection of dark-induced leaf senescence and timing its reversal in barley. Plant Physiol 178:654–671
Steed G, Ramirez DC, Hannah MA, Webb AAR (2021) Chronoculture, harnessing the circadian clock to improve crop yield and sustainability. Science 372:eabc9141. https://doi.org/10.1126/science.abc914
Sugiyama N, Izawa T, Okiawa T, Shimamoto K (2001) Light regulation of circadian clock-controlled gene expression in rice. The Plant J 26:607–615. https://doi.org/10.1046/j.1365-313x.2001.01063.x
Sun L, Dong A, Griffin C, Rongling W (2021) Statistical mechanics of clock gene networks underlying circadian rhythms. Appl Physics Rev 8:021313. https://doi.org/10.1063/5.0029993
Sunoj VSJ, Shroyer KJ, Jagadish SVK, Prasad PVV (2016) Diurnal temperature amplitude alters physiological and growth response of maize (Zea mays L.) during the vegetative stage. Env Exp Bot 130:113–121. https://doi.org/10.1016/j.envexpbot.2016.04.007
Sunoj VSJ, Prasad PVV, Ciampitti IA, Maswada HF (2020) Narrowing diurnal temperature amplitude alters carbon tradeoff and reduces growth in C(4) crop sorghum. Front Plant Sci 11:1262. https://doi.org/10.3389/fpls.2020.01262
Usman B, Nawaz G, Zhao N, Liao S, Liu Y, Li R (2020) Precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa L.) by regulating circadian rhythms and abiotic stress responsive proteins. Int J Mol Sci 21:7854. https://doi.org/10.3390/ijms21217854
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86. https://doi.org/10.1186/s12870-016-0771-y
Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9:491–507. https://doi.org/10.1105/tpc.9.4.491
Wang P, Cui X, Zhao C, Shi L, Zhang G, Sun F, Cao X, Yuan L, **e Q, Xu X (2017) COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J Integr Plant Biol 59:78–85. https://doi.org/10.1111/jipb.12512
Wang F, Han T, Song Q, Ye W, Song X, Chu J, Li J, Chen ZJ (2020a) The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell 32:3124–3138
Wang Y, Yuan L, Su T, Wang Q, Gao Y, Zhang S, Jia Q, Yu G, Fu Y, Cheng Q et al (2020b) Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ 43:637–648. https://doi.org/10.1111/pce.13678
Wang HJ, Ye MX, Fu YR, Dong A, Zhang MM, Feng L, Zhu XL, Bo WH, Jiang LB, Griffin CH et al (2021) Modeling genome-wide by environment interactions through omnigenic interactome networks. Cell Rep 35:109114. https://doi.org/10.1016/j.celrep.2021.109114
Wang P, Wang L, Zhang L, Wu T, Sun B, Zhang J, Sapey E, Yuan S, Jiang B, Chn F et al (2022a) Genomic dissection and diurnal expression analysis reveal the essential roles of the PRR gene family in geographical adaptation of soybean. Int J Mol Sci 23:9970. https://doi.org/10.3390/ijms23179970
Wang Y, Wu F, Lin Q, Sheng P, Wu Z, ** X, Chen W, Li S, Luo S, Duan E et al (2022b) A regulatory loop establishes the link between the circadian clock and abscisic acid signaling in rice. Plant Physiol 191:1857–1870. https://doi.org/10.1093/plphys/kiac548
Webb AAR, Seki M, Satake A, Caldana C (2019) Continuous dynamic adjustment of the plant circadian oscillator. Nat Commun 10:550. https://doi.org/10.1038/s41467-019-08398-5
Wei H, Wang X, He Y, Hang X, Wang L (2021) Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1-mediated sodium homeostasis. EMBO J 40:e105086. https://doi.org/10.15252/embj.2020105086
Wei H, Xu H, Su C, Wang X, Wang L (2022) Rice Circadian Clock Associated 1 transcriptionally regulates ABA signaling to confer multiple abiotic stress tolerance. Plant Physiol 190:1057–1073. https://doi.org/10.1093/plphys/kiac196
Wittern L, Steed G, Taylor LJ, Gardner K, Greenland A, Hannah MA, Webb AAR (2023) Wheat Early Flowering3 is a dawn-expressed circadian oscillator component that regulates heading date. Plant Physiol 191:1383–1403. https://doi.org/10.1093/plphys/kiac544
Wu J-F, Wang Y, Wu S-H (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol 148:948–959. https://doi.org/10.1104/pp.108.124917
Wu J-F, Tsai H-L, Joanito I, Wu Y-C, Chang C-W, Li Y-H, Wang Y, Hong JC, Chu J-W, Hsu C-P et al (2016) LWD–TCP complex activates the morning gene CCA1 in Arabidopsis. Nat Commun 7:13181. https://doi.org/10.1038/ncomms13181
**ang Y, Sapir T, Rouillard P (2022) Interaction between photoperiod and variation in circadian rhythms in tomato. BMC Plant Biol 22:187. https://doi.org/10.1186/s12870-022-03565-1
**e Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, **ng H, Zhi L, Yue Z et al (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26:2843–2857. https://doi.org/10.1105/tpc.114.126573
Xu X, **e Q, McClung CR (2010) Robust circadian rhythms of gene expression in Brassica rapa tissue culture. Plant Physiol 153:841–850. https://doi.org/10.1104/pp.110.155465
Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM (2009) Posttranslational regulation of Circadian Clock Associated1 in the circadian oscillator of Arabidopsis. Plant Physiol 150:844–857. https://doi.org/10.1104/pp.109.137414
Yang L, Liu P, Wang X, Jia A, Ren D, Tang Y, Tang Y, Deng XW, He G (2021) A central circadian oscillator confers defense heterosis in hybrids without growth vigor costs. Nat Commun 12:2317. https://doi.org/10.1038/s41467-021-22268-z
Yu T, Lu X, Bai Y, Mei X, Guo Z, Liu C, Cai Y (2019) Overexpression of the maize transcription factor ZmVQ52 accelerates leaf senescence in Arabidopsis. PLoS ONE 14:e0221949. https://doi.org/10.1371/journal.PONE.0221949
Zhang D, Wang Y, Shen J, Yin J, Li D, Gao Y, Xu W, Liang J (2018a) OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice 11:45. https://doi.org/10.1186/s12284-018-0232-3
Zhang Y, Wang Y, Wei H, Li N, Tian W, Chong K, Wang L (2018b) Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol Plant 11:326–337. https://doi.org/10.1016/j.molp.2017.12.017
Zhang Y-M, Guo P, **a X, Guo H, Zhonghai L (2021) Multiple layers of regulation on leaf senescence: new advances and perspectives. Front Plant Sci 12:788996. https://doi.org/10.3389/fpls.2021.788996
Zhao J, Chen H, Ren D, Tang H, Qiu R, Feng J, Long Y, Niu B, Chen D, Zhong T et al (2015) Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and heading date 3a (Hd3a)/Rice Flowering Locus T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol 208:936–948. https://doi.org/10.1111/nph.13503
Zheng X-M, Feng L, Wang J, Qiao W, Zhang L, Cheng Y, Yang Q (2016) Nonfunctional alleles of long-day suppressor genes independently regulate flowering time. J Integr Plant Biol 58:540–548. https://doi.org/10.1111/jipb.12383
Zheng W-N, Li D-H, Chen F-J, Liu X-P, Li H-Y (2017) Abiotic stress tolerance and ABA responses of transgenic Glycine max plants with modulated RACK1 expression. Can J Plant Sci 99:250–267. https://doi.org/10.1139/cjps-2017-0093
Zou Y, Dai W, Lei W, Su S, Huang Q, Zhou Z, Chen C, Li Z (2018) Identification of proteins interacting with pORF5 in the pathogenesis of C. trachomatis. Am J Transl Res 10:1633–1647
Acknowledgements
CS acknowledges funding support from Science Foundation Ireland (grant 19/FFP/6711). CS and LFQ acknowledge funding support from the Irish Research Council (IRC-GOIPD/2021/710).
Funding
Open Access funding provided by the IReL Consortium. This article is funded by Science Foundation Ireland, 19/FFP/6711, Charles Spillane, Irish Research Council for Science, Engineering and Technology, GOIPD/2021/710, Luis Felipe Quiroz.
Author information
Authors and Affiliations
Contributions
SLD: conceptualization; writing-original draft; writing-review & editing. LFQ and CS: investigation; writing-original draft; writing-review & editing; graphical figure. RW: investigation; writing-original draft; writing-review & editing. AM: investigation; writing-original draft; writing-review & editing. RO: conceptualization; project administration; writing original draft; writing-review & editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests or conflicts of interest, relating to funding, employment or financial (or non-financial) interests. The authors have no relevant financial or non-financial interests to disclose. The authors have no competing interests to declare that are relevant to the content of this article. All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript. The authors have no financial or proprietary interests in any material discussed in this article.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dwivedi, S.L., Quiroz, L.F., Spillane, C. et al. Unlocking allelic variation in circadian clock genes to develop environmentally robust and productive crops. Planta 259, 72 (2024). https://doi.org/10.1007/s00425-023-04324-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04324-8