Abstract
Over the past decade, US Food and Drug Administration (FDA)-approved immune checkpoint inhibitors that target programmed death-1 (PD-1) have demonstrated significant clinical benefit particularly in patients with PD-L1 expressing tumors. Toripalimab is a humanized anti-PD-1 antibody, approved by FDA for first-line treatment of nasopharyngeal carcinoma in combination with chemotherapy. In a post hoc analysis of phase 3 studies, toripalimab in combination with chemotherapy improved overall survival irrespective of PD-L1 status in nasopharyngeal carcinoma (JUPITER-02), advanced non-small cell lung cancer (CHOICE-01) and advanced esophageal squamous cell carcinoma (JUPITER-06). On further characterization, we determined that toripalimab is molecularly and functionally differentiated from pembrolizumab, an anti-PD-1 mAb approved previously for treating a wide spectrum of tumors. Toripalimab, which binds the FG loop of PD-1, has 12-fold higher binding affinity to PD-1 than pembrolizumab and promotes significantly more Th1- and myeloid-derived inflammatory cytokine responses in healthy human PBMCs in vitro. In an ex vivo system employing dissociated tumor cells from treatment naïve non-small cell lung cancer patients, toripalimab induced several unique genes in IFN-γ and immune cell pathways, showed different kinetics of activation and significantly enhanced IFN-γ signature. Additionally, binding of toripalimab to PD-1 induced lower levels of SHP1 and SHP2 recruitment, the negative regulators of T cell activation, in Jurkat T cells ectopically expressing PD-1. Taken together, these data demonstrate that toripalimab is a potent anti-PD-1 antibody with high affinity PD-1 binding, strong functional attributes and demonstrated clinical activity that encourage its continued clinical investigation in several types of cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
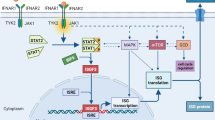
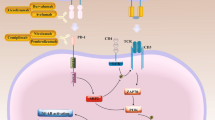
Immune checkpoint inhibitors (ICIs) targeting programmed cell death 1 (PD-1) and programmed cell death ligand 1 (PD-L1) have revolutionized cancer treatment in the recent years, affording long-term survival benefit in a broad range of cancer patients. PD-1 is an inhibitory cell surface receptor that is upregulated upon T cell activation. Upon binding to its ligands PD-L1 and PD-L2, that are expressed on antigen presenting cells and/or tumor cells [1], PD-1 recruits the phosphatases SHP1 and SHP2 which in turn suppresses T cell activation and function [2]. While this mechanism of regulating T cell immune response is necessary in maintaining immune tolerance to autoantigens, several tumors over-express PD-L1 in response to inflammatory mediators and downregulate anti-tumor function of T cells leading to tumor immune evasion [64]. Plots were generated with ggplot2 version 3.4 [65], https://cloud.r-project.org/web/packages/ggplot2/index.html) within R-4.0.2 (https://cran.r-project.org). Due to the analysis of DTC samples generated in two groups, batch correction was performed with combat-seq (Zhang et al., NAR Genomics and Bioinformatics, 2020: 2(3):lqaa078) after iterative testing of gene and sample filtering parameters using principal components analysis with the preprocessCore version 1.56 R package (Bolstad B 2023, https://github.com/bmbolstad/preprocessCore).
Pathway analysis
The logFC values from edgeR differential analysis were used for gene set enrichment analysis (GSEA). GSEA was performed with these gene sets via the clusterProfiler R version 4 package (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html, [66]. The Gene Ontology Biological Process (GOBP) and Hallmark gene sets were obtained from the Broad Institute’s msig.db website (http://www.gsea-msigdb.org/gsea/msigdb). Immune gene sets were extracted from the LM22 matrix [30] by sorting the coefficients for each of the 22 immune cell types and retaining the top 30 genes for each. Immune exhaustion gene sets were derived from the seminal paper on immune exhaustion in the LCMV model system [29].
Statistics
Statistical tests were carried out using GraphPad Prism Software version 9. Statistical tests were used as indicated in the figure legends. p values are reported as follows: *p < 0.05, ** p< 0.01, ***p < 0.001 and ****p < 0.0001.
Data availability
The data supporting the findings in this study are included within the article and its supplementary materials. Raw data that support the findings will be provided upon request.
References
Iwai Y, Hamanishi J, Chamoto K, Honjo T (2017) Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci 24(1):26. https://doi.org/10.1186/s12929-017-0329-9
Arasanz H, Gato-Canas M, Zuazo M, Ibanez-Vea M, Breckpot K, Kochan G et al (2017) PD1 signal transduction pathways in T cells. Oncotarget 8(31):51936–51945. https://doi.org/10.18632/oncotarget.17232
Jiang X, Wang J, Deng X, **ong F, Ge J, **ang B et al (2019) Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer 18(1):10. https://doi.org/10.1186/s12943-018-0928-4
Han Y, Liu D, Li L (2020) PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res 10(3):727–742
Gridelli C, Ardizzoni A, Barberis M, Cappuzzo F, Casaluce F, Danesi R et al (2017) Predictive biomarkers of immunotherapy for non-small cell lung cancer: results from an Experts Panel Meeting of the Italian Association of Thoracic Oncology. Transl Lung Cancer Res 6(3):373–386. https://doi.org/10.21037/tlcr.2017.05.09
Wakabayashi G, Lee YC, Luh F, Kuo CN, Chang WC, Yen Y (2019) Development and clinical applications of cancer immunotherapy against PD-1 signaling pathway. J Biomed Sci 26(1):96. https://doi.org/10.1186/s12929-019-0588-8
Lee JB, Kim HR, Ha SJ (2022) immune checkpoint inhibitors in 10 years: contribution of basic research and clinical application in cancer immunotherapy. Immune Netw 22(1):e2. https://doi.org/10.4110/in.2022.22.e2
Jia L, Zhang Q, Zhang R (2018) PD-1/PD-L1 pathway blockade works as an effective and practical therapy for cancer immunotherapy. Cancer Biol Med 15(2):116–123. https://doi.org/10.20892/j.issn.2095-3941.2017.0086
Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F et al (2018) Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 378(22):2078–2092. https://doi.org/10.1056/NEJMoa1801005
Larkin J, Minor D, D’Angelo S, Neyns B, Smylie M, Miller WH Jr et al (2018) Overall survival in patients with advanced melanoma who received nivolumab versus investigator’s choice chemotherapy in CheckMate 037: a randomized, controlled, open-label phase III trial. J Clin Oncol 36(4):383
Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS et al (2017) Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171(4):934–949. https://doi.org/10.1016/j.cell.2017.09.028
Peng L, Liang WH, Mu DG, Xu S, Hong SD, Stebbing J et al (2021) First-line treatment options for PD-L1-negative non-small cell lung cancer: a bayesian network meta-analysis. Front Oncol 11:657545. https://doi.org/10.3389/fonc.2021.657545
Gogishvili M, Melkadze T, Makharadze T, Giorgadze D, Dvorkin M, Penkov K et al (2022) Cemiplimab plus chemotherapy versus chemotherapy alone in non-small cell lung cancer: a randomized, controlled, double-blind phase 3 trial. Nat Med 28(11):2374–2380. https://doi.org/10.1038/s41591-022-01977-y
Felip E, Altorki N, Zhou C, Vallieres E, Martinez-Marti A, Rittmeyer A et al (2023) Overall survival with adjuvant atezolizumab after chemotherapy in resected stage II-IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase III trial. Ann Oncol 34(10):907–919. https://doi.org/10.1016/j.annonc.2023.07.001
Upadhaya S, Neftelinov ST, Hodge J, Campbell J (2022) Challenges and opportunities in the PD1/PDL1 inhibitor clinical trial landscape. Nat Rev Drug Discov 21(7):482–483
Wang Z, Wu L, Li B, Cheng Y, Li X, Wang X et al (2023) Toripalimab plus chemotherapy for patients with treatment-naive advanced non-small-cell lung cancer: a multicenter randomized phase III trial (CHOICE-01). J Clin Oncol 41(3):651–663. https://doi.org/10.1200/JCO.22.00727
Wang J, Wang Z, Wu L, Li B, Cheng Y, Li X et al (2023) Final overall survival and biomarker analyses of CHOICE-01: a double-blind randomized phase 3 study of toripalimab versus placebo in combination chemotherapy for advanced NSCLC without EGFR/ALK mutations. J Clin Oncol. https://doi.org/10.1200/JCO.2023.41.16_suppl.9003
Wu HX, Pan YQ, He Y, Wang ZX, Guan WL, Chen YX et al (2023) Clinical benefit of first-line programmed death-1 antibody plus chemotherapy in low programmed cell death ligand 1-expressing esophageal squamous cell carcinoma: a post hoc analysis of JUPITER-06 and meta-analysis. J Clin Oncol 41(9):1735–1746. https://doi.org/10.1200/JCO.22.01490
Fuller CL, Braciale VL (1998) Selective induction of CD8+ cytotoxic T lymphocyte effector function by staphylococcus enterotoxin B. J Immunol 161(10):5179–5186
Lee J, Lozano-Ruiz B, Yang FM, Fan DD, Shen L, Gonzalez-Navajas JM (2021) The multifaceted role of Th1, Th9, and Th17 cells in immune checkpoint inhibition therapy. Front Immunol 12:625667. https://doi.org/10.3389/fimmu.2021.625667
Nalbant A (2019) IL-17, IL-21, and IL-22 cytokines of T helper 17 cells in cancer. J Interferon Cytokine Res 39(1):56–60. https://doi.org/10.1089/jir.2018.0057
Voronov E, Dotan S, Krelin Y, Song X, Elkabets M, Carmi Y et al (2013) Unique versus redundant functions of IL-1alpha and IL-1beta in the tumor microenvironment. Front Immunol 4:177. https://doi.org/10.3389/fimmu.2013.00177
Masjedi A, Hashemi V, Hojjat-Farsangi M, Ghalamfarsa G, Azizi G, Yousefi M et al (2018) The significant role of interleukin-6 and its signaling pathway in the immunopathogenesis and treatment of breast cancer. Biomed Pharmacother 108:1415–1424. https://doi.org/10.1016/j.biopha.2018.09.177
Kaneko N, Kurata M, Yamamoto T, Morikawa S, Masumoto J (2019) The role of interleukin-1 in general pathology. Inflamm Regen 39:12. https://doi.org/10.1186/s41232-019-0101-5
Wan J, Wu Y, Ji X, Huang L, Cai W, Su Z et al (2020) IL-9 and IL-9-producing cells in tumor immunity. Cell Commun Signal 18(1):50. https://doi.org/10.1186/s12964-020-00538-5
Suzuki K, Tajima M, Tokumaru Y, Oshiro Y, Nagata S, Kamada H et al (2023) Anti-PD-1 antibodies recognizing the membrane-proximal region are PD-1 agonists that can down-regulate inflammatory diseases. Sci Immunol 8(79):eadd4947. https://doi.org/10.1126/sciimmunol.add4947
Dahan R, Sega E, Engelhardt J, Selby M, Korman AJ, Ravetch JV (2015) FcgammaRs modulate the anti-tumor activity of antibodies targeting the PD-1/PD-L1 axis. Cancer Cell 28(4):543. https://doi.org/10.1016/j.ccell.2015.09.011
Liu H, Guo L, Zhang J, Zhou Y, Zhou J, Yao J et al (2019) Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy. MAbs 11(4):681–690. https://doi.org/10.1080/19420862.2019.1596513
Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V et al (2007) Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27(4):670–684. https://doi.org/10.1016/j.immuni.2007.09.006
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259. https://doi.org/10.1007/978-1-4939-7493-1_12
Zhou Y, Goenaga AL, Harms BD, Zou H, Lou J, Conrad F et al (2012) Impact of intrinsic affinity on functional binding and biological activity of EGFR antibodies. Mol Cancer Ther 11(7):1467–1476. https://doi.org/10.1158/1535-7163.MCT-11-1038
Chen D, Tan S, Zhang H, Wang H, He W, Shi R et al (2019) The FG loop of PD-1 serves as a “Hotspot” for therapeutic monoclonal antibodies in tumor immune checkpoint therapy. iScience 14:113–24. https://doi.org/10.1016/j.isci.2019.03.017
Yokosuka T, Saito T (2010) The immunological synapse, TCR microclusters, and T cell activation. Curr Top Microbiol Immunol 340:81–107. https://doi.org/10.1007/978-3-642-03858-7_5
Pentcheva-Hoang T, Chen L, Pardoll DM, Allison JP (2007) Programmed death-1 concentration at the immunological synapse is determined by ligand affinity and availability. Proc Natl Acad Sci U S A 104(45):17765–17770. https://doi.org/10.1073/pnas.0708767104
Elia AR, Grioni M, Basso V, Curnis F, Freschi M, Corti A et al (2018) Targeting tumor vasculature with TNF leads effector T cells to the tumor and enhances therapeutic efficacy of immune checkpoint blockers in combination with adoptive cell therapy. Clin Cancer Res 24(9):2171–2181. https://doi.org/10.1158/1078-0432.CCR-17-2210
Wei SC, Anang NAS, Sharma R, Andrews MC, Reuben A, Levine JH et al (2019) Combination anti-CTLA-4 plus anti-PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies. Proc Natl Acad Sci U S A 116(45):22699–22709. https://doi.org/10.1073/pnas.1821218116
Gubin MM, Esaulova E, Ward JP, Malkova ON, Runci D, Wong P et al (2018) High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell 175(4):1014–1030. https://doi.org/10.1016/j.cell.2018.09.030
Harper TA, Bacot SM, Fennell CJ, Matthews RL, Zhu C, Yue P et al (2021) IL-10 signaling elicited by Nivolumab-induced activation of the MAP kinase pathway does not fully contribute to Nivolumab-modulated heterogeneous T cell responses. Int J Mol Sci. https://doi.org/10.3390/ijms222111848
Yang AS, Lattime EC (2003) Tumor-induced interleukin 10 suppresses the ability of splenic dendritic cells to stimulate CD4 and CD8 T-cell responses. Cancer Res 63(9):2150–2157
Sullivan KM, Jiang X, Guha P, Lausted C, Carter JA, Hsu C et al (2023) Blockade of interleukin 10 potentiates antitumour immune function in human colorectal cancer liver metastases. Gut 72(2):325–337. https://doi.org/10.1136/gutjnl-2021-325808
Wang Q, Bardhan K, Boussiotis VA, Patsoukis N (2021) The PD-1 interactome. Adv Biol (Weinh) 5(9):e2100758. https://doi.org/10.1002/adbi.202100758
Zheng Y, Han L, Chen Z, Li Y, Zhou B, Hu R et al (2022) PD-L1(+)CD8(+) T cells enrichment in lung cancer exerted regulatory function and tumor-promoting tolerance. iScience 25(2):103785. https://doi.org/10.1016/j.isci.2022.103785
Mazet JM, Mahale JN, Tong O, Watson RA, Lechuga-Vieco AV, Pirgova G et al (2023) IFNgamma signaling in cytotoxic T cells restricts anti-tumor responses by inhibiting the maintenance and diversity of intra-tumoral stem-like T cells. Nat Commun 14(1):321. https://doi.org/10.1038/s41467-023-35948-9
Strauss L, Mahmoud MAA, Weaver JD, Tijaro-Ovalle NM, Christofides A, Wang Q et al (2020) Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci Immunol. https://doi.org/10.1126/sciimmunol.aay1863
Hulme EC, Trevethick MA (2010) Ligand binding assays at equilibrium: validation and interpretation. Br J Pharmacol 161(6):1219–1237. https://doi.org/10.1111/j.1476-5381.2009.00604.x
Yu X, Orr CM, Chan HTC, James S, Penfold CA, Kim J et al (2023) Reducing affinity as a strategy to boost immunomodulatory antibody agonism. Nature 614(7948):539–547. https://doi.org/10.1038/s41586-022-05673-2
Yu X, Chan HTC, Orr CM, Dadas O, Booth SG, Dahal LN et al (2018) Complex interplay between epitope specificity and isotype dictates the biological activity of anti-human CD40 antibodies. Cancer Cell 33(4):664–675. https://doi.org/10.1016/j.ccell.2018.02.009
Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N (2016) High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep 6:35297. https://doi.org/10.1038/srep35297
Richaud AD, Zaghouani M, Zhao G, Wangpaichitr M, Savaraj N, Roche SP (2022) Exploiting the innate plasticity of the programmed cell death-1 (PD1) receptor to design pembrolizumab H3 loop mimics. ChemBioChem 23(21):e202200449. https://doi.org/10.1002/cbic.202200449
Mai H-Q, Chen Q-Y, Chen D, Hu C, Yang K, Wen J et al (2021) Toripalimab or placebo plus chemotherapy as first-line treatment in advanced nasopharyngeal carcinoma: a multicenter randomized phase 3 trial. Nat Med 27(9):1536–1543
Wang Z-X, Cui C, Yao J, Zhang Y, Li M, Feng J et al (2022) Toripalimab plus chemotherapy in treatment-naïve, advanced esophageal squamous cell carcinoma (JUPITER-06): a multi-center phase 3 trial. Cancer Cell 40(3):277–288
Wang Z, Wu L, Li B, Cheng Y, Li X, Wang X et al (2023) toripalimab plus chemotherapy for patients with treatment-naive advanced non–small-cell lung cancer: a multicenter randomized phase III trial (CHOICE-01). J Clin Oncol 41(3):651
Mai H-Q, Chen Q-Y, Chen D, Hu C, Yang K, Wen J et al (2023) Toripalimab plus chemotherapy for recurrent or metastatic nasopharyngeal carcinoma: the JUPITER-02 randomized clinical trial. JAMA 330(20):1961–1970. https://doi.org/10.1001/jama.2023.20181
Noori M, Yousefi AM, Zali MR, Bashash D (2022) Predictive value of PD-L1 expression in response to immune checkpoint inhibitors for esophageal cancer treatment: a systematic review and meta-analysis. Front Oncol 12:1021859. https://doi.org/10.3389/fonc.2022.1021859
Chau I, Doki Y, Ajani JA, Xu J, Wyrwicz L, Motoyama S et al (2021) Nivolumab (NIVO) plus ipilimumab (IPI) or NIVO plus chemotherapy (chemo) versus chemo as first-line (1L) treatment for advanced esophageal squamous cell carcinoma (ESCC): first results of the CheckMate 648 study. J Clin Oncol 39:LBA4001-LBA. https://doi.org/10.1200/JCO.2021.39.15_suppl.LBA4001
Sun J-M, Shen L, Shah MA, Enzinger P, Adenis A, Doi T et al (2021) Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): a randomised, placebo-controlled, phase 3 study. Lancet 398(10302):759–71
Luo H, Lu J, Bai Y, Mao T, Wang J, Fan Q et al (2021) Effect of camrelizumab vs placebo added to chemotherapy on survival and progression-free survival in patients with advanced or metastatic esophageal squamous cell carcinoma: the ESCORT-1st randomized clinical trial. JAMA 326(10):916–925. https://doi.org/10.1001/jama.2021.12836
Sheng X, Chen H, Hu B, Yao X, Liu Z, Yao X et al (2022) Safety, efficacy, and biomarker analysis of toripalimab in patients with previously treated advanced urothelial carcinoma: results from a multicenter phase II trial POLARIS-03. Clin Cancer Res 28(3):489–497. https://doi.org/10.1158/1078-0432.CCR-21-2210
Emens LA, Middleton G (2015) The interplay of immunotherapy and chemotherapy: harnessing potential synergies. Cancer Immunol Res 3(5):436–443. https://doi.org/10.1158/2326-6066.CIR-15-0064
Wang ZX, Cui C, Yao J, Zhang Y, Li M, Feng J et al (2022) Toripalimab plus chemotherapy in treatment-naive, advanced esophageal squamous cell carcinoma (JUPITER-06): a multi-center phase 3 trial. Cancer Cell 40(3):277–28. https://doi.org/10.1016/j.ccell.2022.02.007
Mai HQ, Chen QY, Chen D, Hu C, Yang K, Wen J et al (2021) Toripalimab or placebo plus chemotherapy as first-line treatment in advanced nasopharyngeal carcinoma: a multicenter randomized phase 3 trial. Nat Med 27(9):1536–1543. https://doi.org/10.1038/s41591-021-01444-0
Mai H-Q, Chen Q-Y, Chen D, Hu C, Yang K, Wen J et al (2022) Abstract CT226: Final progression-free survival analysis of JUPITER-02, a randomized, double-blind, phase 3 study of toripalimab or placebo plus gemcitabine and cisplatin as first-line treatment for recurrent or metastatic nasopharyngeal carcinoma. Cancer Res 82:CT226-CT
Wang Z, Ying J, Xu J, Yuan P, Duan J, Bai H et al (2020) Safety, antitumor activity, and pharmacokinetics of toripalimab, a programmed cell death 1 inhibitor, in patients with advanced non-small cell lung cancer: a phase 1 trial. JAMA Netw Open 3(10):e2013770. https://doi.org/10.1001/jamanetworkopen.2020.13770
Pol E, Roos H, Markey F, Elwinger F, Shaw A, Karlsson R (2016) Evaluation of calibration-free concentration analysis provided by Biacore systems. Anal Biochem 510:88–97. https://doi.org/10.1016/j.ab.2016.07.009
Wickham H (2009) Ggplot2: elegant graphics for data analysis. Springer New York, Dordrecht, New York
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2(3):100141. https://doi.org/10.1016/j.xinn.2021.100141
Acknowledgements
We thank the patients who volunteered to participate in the toripalimab clinical studies and their families, all the investigators and study site personnel. We thank Marc Pondel, Rosh Dias, Nathalie Vandenkoornhuyse-Yanze and the Coherus Scientific Advisory Board members for thoughtful review and editorial input into the manuscript.
Author information
Authors and Affiliations
Contributions
N. R: Conceptualization, data curation, formal analysis, investigation, writing–original draft, project administration, writing– review and editing. X.W: Investigation, data curation, formal analysis, writing–original draft, writing– review and editing. S. R: Investigation, data curation, formal analysis, writing–original draft, writing– review and editing. D. C: Investigation, Data curation, formal analysis, writing–original draft, writing– review and editing. Sy.T: Investigation, Data curation, formal analysis, writing–original draft, writing– review and editing. S.K: Investigation, Data curation, formal analysis, writing–original draft, writing– review and editing. K.W: Investigation, Data curation, formal analysis, writing–original draft, writing– review and editing. V. K: Investigation, Data curation, formal analysis, writing–original draft, writing– review and editing. V. V: Writing–original draft, writing– review and editing. P. K: Investigation, formal analysis, writing–original draft, writing– review and editing. S.Y: Investigation, formal analysis, writing–original draft, writing– review and editing. T.L: Conceptualization, writing–original draft, project administration, writing– review and editing. S.K: Conceptualization, writing–original draft, project administration, writing– review and editing.
Corresponding author
Ethics declarations
Competing interests
NR, XW, SR, DC, ST and SK report employment and stock ownership with Coherus Biosciences, Inc. TL reports Coherus Biosciences, Inc. employment and Coherus Biosciences and AstraZeneca stock ownership. PK reports employment with TopAlliance BioSciences Inc. SY reports employment with TopAlliance Biosciences Inc. and Shanghai Junshi Biosciences and stock ownership with Shanghai Junshi Biosciences. VV and SDK report stock ownership with Coherus Biosciences.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rajasekaran, N., Wang, X., Ravindranathan, S. et al. Toripalimab, a therapeutic monoclonal anti-PD-1 antibody with high binding affinity to PD-1 and enhanced potency to activate human T cells. Cancer Immunol Immunother 73, 60 (2024). https://doi.org/10.1007/s00262-024-03635-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00262-024-03635-3