Abstract
The restricted diversity of the major histocompatibility complex (MHC) of Mauritian cynomolgus macaques provides powerful opportunities for insight into host-viral interactions and cellular immune responses that restrict lentiviral infections. However, little is known about the effects of Mhc haplotypes on control of SIV in this species. Using microsatellite-based genoty** and allele-specific PCR, Mhc haplotypes were deduced for 35 macaques infected with the same stock of SIVmac251. Class I haplotype H6 was associated with a reduction in chronic phase viraemia (p = 0.0145) while a similar association was observed for H6 class II (p = 0.0063). An increase in chronic phase viraemia, albeit an insignificant trend, was observed in haplotype H5-positive animals. These results further emphasise the value of genetically defined populations of non-human primates in AIDS research and provide a foundation for detailed characterisation of MHC restricted cellular immune responses and the effects of host genetics on SIV replication in cynomolgus macaques.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infection of cynomolgus macaques (Macaca fascicularis) with SIV represents a frequently used model of HIV/AIDS. This macaque species has been employed in studies of pathogenesis (Dittmer et al. 1996; Feichtinger et al. 1990; Habis et al. 1999; Maggiorella et al. 1998; Montgomery et al. 1999; Putkonen et al. 1989), evaluation of antiretroviral therapy (Brandin et al. 2006; Jorajuria et al. 2000; Le Grand et al. 1994; Tsai et al. 1998) and vaccine development (Berry et al. 2007; Gotch et al. 1991; Maggiorella et al. 2007; Negri et al. 2004; Nilsson et al. 2001; Putkonen et al. 1998; Stebbings et al. 2004; Titti et al. 1997; Wade-Evans et al. 2001). Central to the development of a broadly cross-reactive HIV vaccine is an understanding of the factors that determine the specificity of anti-HIV responses, particularly in the case of T-cell vaccines designed to elicit cellular immune responses. In order to fully characterise such responses in an animal model, knowledge of the Mhc allele repertoire of the animals used, and the impact of these alleles on viral dynamics, is essential.
Certain HLA molecules have been associated with effective control of HIV, notably HLA-B27 and HLA-B57 (Kaslow et al. 1996; McNeil et al. 1996; Migueles et al. 2000), and these restrict potent anti-HIV cytotoxic T-lymphocyte (CTL) responses (Gillespie et al. 2002; Migueles et al. 2000; Nelson et al. 1997). Similarly, MHC molecules encoded by rhesus macaque Mhc class I alleles Mamu-A*01, Mamu-B*08 and Mamu-B*17 have been reported to present SIV-derived peptides that elicit potent cellular immune responses, and are associated with lower chronic phase viral loads (Loffredo et al. 2007a, b; Mothe et al. 2003; Muhl et al. 2002; O'Connor et al. 2003; Yant et al. 2006; Zhang et al. 2002). It is essential to control for such host bias by appropriate distribution of animals carrying these alleles among experimental groups, to prevent incorrect interpretation of data. However, an understanding of how particular Mhc genotypes confer superior control of viral replication will provide valuable information about the induction of potent cellular immune responses generated following infection, and identify how these responses might be harnessed by vaccination.
Limited data are available on the effect of Mhc haplotypes of M. fascicularis on control of SIV infection in Mauritian cynomolgus macaques (MCM). One study demonstrated that MCM with identical Mhc haplotypes mounted comparable cellular immune responses and maintained similar viral loads following infection with the widely used virus isolate SIVmac239 (Wiseman et al. 2007). Further, H3 and H6 class IB and class II haplotypes were associated with resistance to chimaeric SHIV89.6P challenge, while H2 and H5 class IB and class II haplotypes were associated with susceptibility to infection (Florese et al. 2008). To date, however, no study has described the effect of Mhc haplotypes on control of SIVmac replication in a large cohort of genetically defined cynomolgus macaques.
Here we describe the first systematic evaluation of the effect of Mhc class I haplotypes on SIV replication in a cohort of 35 MCM infected with an identical stock of pathogenic SIVmac251. Using microsatellite analysis and novel allele-specific PCR, we deduced Mhc haplotypes for all animals in the cohort. Haplotype H6 was associated significantly with a reduction in chronic phase viral load while animals positive for haplotype H5 failed to control viraemia as well as other animals in the cohort. These findings have important implications for design of experiments using MCM, and provide a foundation for further detailed analysis of anti-SIV cellular immune responses elicited in this species.
Materials and methods
Animals
Biological material and virological data from 35 Mauritian cynomolgus macaques challenged with SIVmac251 as part of an EU-funded pre-clinical vaccine evaluation and 14 animals used in virus titration experiments were available for the current study. The 49 samples were obtained from three centres located in France, Italy and the UK that used common batches of recombinant SIV vaccines in different regimens prior to intra-rectal challenge with an identical stock of SIVmac251. Thirty-five samples were included in the analysis and the remaining 14 were excluded due to failure to become infected (six vaccinees were protected against infection, i.e. no or low transient viraemia; one control failed to become infected for technical reasons; seven titration animals failed to become infected). Vaccination and infectious challenge regimens and exclusion criteria are described in Table 1 of the Electronic supplementary material. Animals were challenged intra-rectally with an identical stock of SIVmac251 RKI (Neildez et al. 1998). Animals were housed and maintained in accordance with appropriate national guidelines for care and maintenance of non-human primates that fulfil EEC Directive No. 86-609, 24 November 1986.
vRNA quantification
Plasma viral loads were quantified by quantitative real-time RT-PCR assays, each with a sensitivity of 50 copies/ml as previously described (Berry et al. 2008; Negri et al. 2004; ten Haaft et al. 1998, 2001). The comparability of data for assays performed at different centres has been evaluated using common reference materials and variation determined to be less than 5% (N.B. unpublished data).
DNA extraction
EDTA- or citrate-treated blood samples were obtained from all animals. DNA was isolated by phenol–chloroform extraction, resuspended in molecular-grade water, quantified using Hoescht dye (Sigma-Aldrich, Dorset, UK) and diluted to 20 ng/μl in molecular grade water.
Definition of Mhc haplotypes
Mhc haplotypes of animals were determined by microsatellite PCR as described previously (Mee et al. 2009). Where recombination was observed in the Mhc class I region, allele-specific PCR was employed to resolve the individual alleles carried by each animal. For analysis purposes, if a recombinant haplotype was found to carry all class I alleles associated with a particular haplotype, the animal was scored positive for that class I haplotype.
Allele-specific PCR
Allele-specific PCR primers were designed against polymorphic regions of Mhc class I heavy chain genes described previously in Mauritian cynomolgus macaques (Wiseman et al. 2007). Genomic DNA (60 ng) was amplified in a 15 μl reaction comprising 1× PCR Reaction Buffer with MgCl2 (final concentration 2 mM MgCl2 unless specified otherwise), 1× GC-Rich solution, 200 μM each dNTPs, 0.55 U FastStart Taq (all Roche Diagnostics, Burgess Hill, UK) and allele-specific primers (Invitrogen, Paisley, UK). Primers designed to amplify a ∼300-bp product from the DRB locus, DRB-N (5′-GCC TCG AGT GTC CCC CCA GCA CGT TTC-3′) and DRB-C (5′-GCC GCA GCT TTC ACC TCG CCG CTG-3′) were included as an internal control. Primer sequences and amplification conditions for each Mhc class I allele are described in Table 1. Cycling conditions consisted of an initial denaturation at 95°C for 5 min, n cycles (see Table 1) of denaturation at 94°C for 30 s, annealing at the indicated temperature for 30 s and extension at 72°C for 40 s, followed by a final extension at 72°C for 5 min. Amplified products were separated by electrophoresis through 1.7% agarose (Invitrogen) at 150 V for 1.5 h and visualised by ethidium bromide staining and exposure to ultraviolet light. Bands of the expected size, relative to a ΦX174/HaeIII size ladder (Promega, Southampton, UK) were excised from the agarose gel and purified using High Pure PCR cleanup micro kit (Roche Diagnostics) and bi-directionally sequenced with modified M13 forward (5′-AAA ACG ACG GCC AGT-3′) and reverse (5′-GAA ACA GCT ATG ACC-3′) primers (Invitrogen) and BigDye Terminator v3.1 cycle sequencing (Applied Biosystems, Cheshire, UK). Electrophoresis and automated base calling were performed on an ABI3130xl genetic analyzer (Applied Biosystems) and contiguous sequences were assembled using Trace Edit Pro (Ridom GmbH, Würzburg, Germany).
Statistical analysis
Statistical analysis was performed using Minitab 15 (Minitab Inc., PA, USA). All viral loads were log transformed prior to analysis. For each animal, chronic phase viral load was calculated as the mean of all measurements from week 12 post-infection to termination of experiment or 52 weeks. Groups of animals were segregated on the basis of Mhc haplotypes and mean viral loads were compared between groups using a two-tailed unpaired t test. Viral loads were compared for all animals with and without each haplotype. Results are presented with and without application of the Bonferroni correction for multiple comparisons.
Results
Allele-specific PCR for Mhc-B alleles of Mauritian cynomolgus macaques
A panel of 20 novel allele-specific PCR primer sets was employed to successfully amplify the majority of Mhc-B alleles expressed in Mauritian cynomolgus macaques. Specificity of these assays was assessed against eight samples from the UK MCM breeding facility representing all seven MCM Mhc haplotypes (data not shown). Sequencing of all amplified products confirmed that, with the exception of primers for Mafa-B*460101 which cross-reacted with B*480101, all primer sets were highly specific under the described reaction conditions (Table 1). Allele-specific PCR was employed to resolve eight recombinant haplotypes in seven animals in the study cohort, with confirmation of amplicon identity by sequencing. Sequences of all amplicons have been deposited in the EMBL Nucleotide Sequence Database and accession numbers are provided in Table 1.
Characterisation of Mhc haplotypes
All animals in the study cohort carried only the seven common Mhc haplotypes (H1–H7) found in Mauritian cynomolgus macaques, or simple recombinants thereof (Fig. 1a, b). Resolution of recombinant class IA and class II haplotypes was not attempted as such recombination was observed in only three animals in each case. Furthermore, in the case of the class IA region, the recombination did not alter the class IA allele composition since several alleles are shared by the three most common class IA haplotypes (Wiseman et al. 2007). Where recombination was observed within the class IB region, allele-specific PCR was employed to resolve the alleles carried by each animal. With the exception of animal X295 (which inherited Mafa-B*490101, B*650101 and I*100201, but not B*640101 from the H6 haplotype, and inherited no alleles from the H1 haplotype) and animal 1980 (which inherited B*430101 but not B*440101 or B*460101 from H1) recombination in the class IB region resulted in the inheritance of alleles from only one of the parent haplotypes (Fig. 1a, b). Only animals inheriting all class IB alleles associated with a given haplotype were considered positive for this class IB haplotype for subsequent analysis. Class I haplotype H1 was the most frequent haplotype in the cohort (Fig. 1c).
a Mhc haplotypes of animals infected with SIVmac251. Mhc haplotypes were determined by microsatellite analysis and haplotypes with recombination within the class IB region were resolved by allele-specific PCR. Intact haplotypes are indicated underneath the data for each animal. H1–H7 intact haplotypes previously identified in Mauritian cynomolgus macaques (Mee et al. 2009; Wiseman et al. 2007). rec recombinant haplotype; Vacc. vaccine regimen as detailed in Table 1 of the Electronic supplementary material; Vect. vector regimen; Titr. naïve animal used in titration experiment; naïve naïve challenge control. b Mhc haplotypes of vaccinated animals excluded from analysis. c Percentage of infected animals in study cohort carrying each Mhc class I haplotype
Impact of Mhc haplotype on virological outcome of challenge
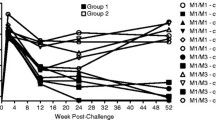
When grouped according to Mhc class I haplotype, differences were observed in chronic phase viral load (Fig. 2). Animals positive for haplotype H6 had lower viral loads than the mean for all animals (Figs. 2 and 3a, b). One H6-positive animal (62955) suppressed viraemia to undetectable levels from week 18 and remained aviraemic until termination of the study at 97 weeks (Fig. 3b and data not shown). Conversely, the viral load in the two animals positive for H5 was higher than in H5-negative animals at all time points from week 4 onwards, with the difference becoming more pronounced towards the end of the study period (Figs. 2 and 3c, d). A trend towards lower chronic phase viral load was also noted in animals positive for haplotype H7 (Figs. 2 and 3e, f).
Viral load in animals carrying specific Mhc class I haplotypes. Viral RNA was quantified by quantitative real-time RT-PCR at the indicated time points. Data represent the geometric mean viral load of all animals carrying an intact copy of the indicated Mhc class I haplotype. Dotted line represents assay cutoff (50 vRNA copies/ml). Error bars are omitted for clarity
Mhc class I haplotype H6 (H6-I) is associated with lower chronic phase viral loads. a Geometric mean viral load for all H6-I-positive (n = 3) and H6-I-negative (n = 32) animals; b individual viral loads of H6-positive animals; c geometric mean viral load for all H5-positive (n = 2) and H5-negative (n = 33) animals; d individual viral loads of H5-positive animals; e geometric mean viral load for all H7-positive (n = 2) and H7-negative (n = 33) animals; f individual viral loads of H7-positive animals. Broken lines represent animals positive for either the indicated class IA or B haplotype, but not both. Dotted line represents assay cutoff (50 vRNA copies/ml). Error bars denote 95% confidence intervals
In order to quantify the difference in viral load conferred by specific haplotypes, the chronic phase viral load (defined as the geometric mean of all viral load measurements between week 12 and termination of study) was compared between animals positive and negative for each class I haplotype. Mhc class I haplotype H6 (H6-I) was associated with a significant reduction in chronic phase viral load of 1.92 log10 vRNA copies/ml from the mean (Fig. 4a, p = 0.0145). The H6-I association remained significant if all vaccinated animals were excluded from the analysis (p = 0.0214, data not shown). Animals positive for Mhc class I haplotype H5 (H5-I) had a relative increase of 1.52 log10 viral RNA copies/ml in the chronic phase; however, this did not reach statistical significance (Fig. 4a). Two animals in the study were positive for haplotype H7. These animals had lower chronic phase viral loads, comparable to H6-I-positive animals, but this observation did not reach statistical significance (Figs. 2 and 4).
Relative viral load difference associated with each Mhc haplotype. a Relative difference in chronic phase viral load (geometric mean of all measurements between weeks 12 and 53 or termination of experiment) associated with class I haplotypes (i.e. class IA and class IB region belonging to the same haplotype); b relative difference in chronic phase viral load associated with class IA haplotypes; c relative difference in chronic phase viral load associated with class IB haplotypes; d relative difference in chronic phase viral load associated with class II haplotypes. Only animals with an intact copy of the indicated Mhc region are included in the analysis. Circles indicate mean difference in viral load in all animals positive for indicated haplotypes relative to those lacking the indicated haplotype. p values indicate significant results as determined by Student’s t test. Error bars denote 95% confidence intervals
Since several animals in the cohort carried simple recombinant haplotypes, we also grouped animals according to class IA, IB or class II haplotype. No association was observed between class IA haplotypes and altered viral load in this cohort of animals (Fig. 4b). A significant association was noted between Mhc class IB haplotype H6 (H6-IB, p = 0.0282) and Mhc class II haplotype H6 (H6-II, p = 0.0063) regions and lower viral load (Fig. 4c, d). Both these findings remained statistically significant if vaccinated animals were excluded from the analysis (H6-IB, p = 0.0411; H6-II, p = 0.0096; data not shown).
A more stringent analysis of the above results revealed that the associations for H6-I and H6-IB did not reach statistical significance following application of the Bonferroni correction for multiple comparisons (corrected significance level for seven tests—p < 0.0071). The association between H6 class II and low viral loads remained significant following correction (p = 0.0063).
Effect of Mhc haplotype combinations on viral replication
Three animals in the cohort, including the only animal that failed to be protected by vaccine regimen 2, shared identical Mhc class I haplotypes, being heterozygous for H1 and H4. We had previously observed this combination of Mhc class I haplotypes in a number of animals that failed to control infection with a variety of SIV isolates (E.T.M., N.B., C.H., N.A., N.J.R., unpublished observations). The mean viral load for animals carrying H1 and H4 was approximately 30-fold higher than all other haplotypes and tended towards, but did not reach, statistical significance (p = 0.0573, data not shown).
Effect of Mhc haplotypes on vaccination outcome
Six of 15 vaccinated macaques were protected against challenge with SIVmac251 by vaccination (0/4 with Vaccine 1, 3/4 with Vaccine 2, 3/5 with Vaccine 3) as defined by transient or undetectable viraemia during the follow-up period (28 weeks for Vaccine 1, >52 weeks for Vaccines 2 and 3, data not shown). Taking the combined vaccine studies together, there was no significant bias in haplotype frequency in the protected animals compared to unprotected vaccinated animals or naïve and vector controls (Fig. 1b). None of the vaccinated animals carried haplotype H6. Therefore, Mhc haplotypes were not associated with vaccination outcome in this study.
Discussion
The identification of a limited number of high frequency Mhc haplotypes in Mauritian cynomolgus macaques (Wiseman et al. 2007) established the exceptional potential of this population of non-human primates in biomedical research. In particular, it facilitates detailed analysis of the impact of immunogenetics in non-human primate models of infectious disease and provides a step** stone for the development of new reagents to characterise cellular immunity following vaccination or infection, particularly for studies of cellular immunity in the context of HIV pathogenesis and vaccine evaluation. In this retrospective analysis of an EU-funded collaborative research programme into candidate AIDS vaccine strategies, we have investigated whether Mhc genetics influence chronic phase viral load in SIVmac251 infection of Mauritian cynomolgus macaques.
We have employed microsatellite-based genoty** to ascertain the Mhc haplotypes of 35 Mauritian cynomolgus macaques infected with the same stock of SIVmac251. In addition, to facilitate accurate genoty** of animals with recombinant haplotypes, we developed a panel of allele-specific PCR primers to amplify the majority of Mhc class IB alleles in MCM. These reagents will be of use to researchers beyond the scope of the current study. In the largest comparative analysis performed to date, we studied the impact of Mhc haplotypes on the control of viral replication in the chronic phase of infection. There was a statistically significant association between H6 class I and reduced chronic phase viraemia, while animals with haplotype H5 had higher than average viral loads and those with H7 had lower viral loads, though it should be noted that only two animals were available for each of haplotypes H5 and H7. A further observation was that animals with the Mhc class I haplotype combination H1–H4 had higher chronic phase viral loads, though these latter results did not reach statistical significance.
Through analysis of simple recombinant Mhc haplotypes, the protective effect of haplotype H6-I was associated with the class IB region, but not the A region. This would suggest that the observed effect is mediated by gene products of the class IB region, consistent with the reported greater role of MHC-B alleles in mediating effective control of HIV in humans (Bihl et al. 2006; Kiepiela et al. 2004) and SIV in macaques (Loffredo et al. 2007a, b; O'Connor et al. 2003; Yant et al. 2006). It should be noted that, in three animals, the H6 class I haplotype extended across both the class IA and class IB regions. Analysis of further animals positive for either H6 class IA or H6 class IB, but not both, would establish conclusively whether the observed association is the result of the indicated Mhc region or of the extended Mhc haplotype. However, the low frequency of haplotype H6 among MCM means that the retrospective identification of sufficient numbers of such animals will be difficult.
Since the 35 animals analysed were drawn from groups that had received candidate SIV vaccines, as well as those receiving empty vaccine vectors and naïve challenge controls, there is a possibility that these data could have been obtained due to unequal randomisation of haplotypes between groups. Indeed, six vaccinated animals from the original cohort were excluded from the analysis due to failure to become infected. Following exclusion of these animals from analysis, no differences in chronic phase viral load were observed between groups receiving different treatments. Notably all of the H6 class I- and H6 class IB-positive animals in this study were naïve challenge controls. Additionally, no significant difference in peak or chronic phase viral load was observed between groups of animals receiving different challenge doses. Hence, bias due to prior vaccination or differences in challenge dose can be excluded in this situation. Furthermore, no association was noted between MHC haplotype and vaccine-induced protection against challenge.
Our findings are consistent with a previous study describing superior and inferior control of SHIV89.6Pcy243 replication by animals carrying haplotypes H6 and H5, respectively (Florese et al. 2008). In contrast, we did not observe a beneficial effect of haplotype H3, nor a detrimental effect of H2 as reported by Florese and co-workers, possibly due to the use of a cynomolgus macaque-adapted SHIV expressing HIV-derived env, tat, rev and vpu genes (Borsetti et al. 2008) in that study, as opposed to unmodified SIV in the present study. Observations in a limited number of animals suggest that H6-positive animals also control replication of other highly pathogenic strains of SIVmac251 (E.T.M., N.B., C.H., N.A., N.J.R., unpublished observations). We have initiated further retrospective analyses using material from cohorts of animals infected with a range of SIV stocks in addition to animals protected against pathogenic SIV infection by immunisation with live-attenuated virus (Almond and Stott 1999; Berry et al. 2008; Stebbings et al. 2002, 2004) to investigate whether the beneficial effect of haplotype H6 may be broadly applicable to a variety of SIV/SHIV stocks.
The association found between the H6 class II haplotype and reduced viral load was unexpected. The vast majority of previous studies have focused on associations between MHC class I alleles and lentiviral infection. Nevertheless, a limited number of reports have suggested an association between MHC class II alleles and control of SIV (Giraldo-Vela et al. 2008) and HIV (Hardie et al. 2008a, b; Lacap et al. 2008; Ndung'u et al. 2005) and we have previously noted a protective effect of Mafa-DRB1*0307 in relation to infection with simian retrovirus, type 2 (Mee et al. 2008). Though the numbers of H6-II-positive animals were low (n = 3), in two of the animals the H6 haplotype did not extend across the class I region (Fig. 1). It is possible therefore that the class IB region and class II region of H6 mediate protective effects independently of one another. The class II region of MCM thus merits further investigation.
A caveat to the above findings is that following a stricter analysis of the data, with correction for multiple testing, the H6 class I and H6 class IB associations did not reach statistical significance. This reflects the difficulties in identifying large numbers of animals positive for a low-frequency haplotype in studies where group sizes are necessarily limited by ethical constraints. Nevertheless, our data represent an important development in the use of MCM in AIDS vaccine research and emphasise the importance of cohort studies such as this where data can be drawn from a number of retrospective studies to increase statistical power.
Our results and those of a previous study (Florese et al. 2008) underscore the importance of characterisation of host genetics prior to selection of cynomolgus macaques for HIV vaccine evaluation. Macaques positive for H6 should either be excluded from vaccine studies or distributed evenly among experimental groups to avoid introducing bias. Likewise, though the differential viral loads noted for haplotypes H5 and H7 did not reach statistical significance, these animals should be distributed evenly among experimental groups until the phenotypes associated with each haplotype can be conclusively defined.
The finding that haplotype H6 is associated with superior control of the chronic phase of SIV replication will permit further investigation of the mechanism by which this phenotype is mediated. Previous identification of Mhc alleles linked to better control of HIV or SIV have focused on MHC class I restriction of cytotoxic T cell responses, though the identification of similar superior control of viral replication in animals positive only for the class II region of H6 indicates that MHC class II restricted CD4+ T cell responses may also contribute to the observed phenotype. Map** of CTL epitopes and their restricting class I alleles in animals with protective haplotypes is required to complement genetic studies, and will establish whether classical MHC-restricted CTL activity is responsible for the observed protection or whether it is the result of linked genetic loci. To address these issues, we have initiated development of reagents for the study of cellular immunity in MCM, including cell lines expressing individual MHC-I alleles and B- and T-cell lines for peptide map** and identification of specific CTL in infected and/or immunised animals. We have also extended our established selective breeding programme (Mee et al. 2009) to increase the frequency of animals with haplotypes H4–H7 to facilitate further study.
In most situations studied to date, MHC association with a disease phenotype is a result of differences in adaptive immune responses restricted by a specific allele. The possibility exists, however, that genes other than those encoding classical MHC antigens are important, or that classical MHC gene products may be mediating control indirectly. For example, Natural Killer Cell Immunoglobulin-like Receptors (NK-KIR) and their primary ligands, MHC-I molecules, can mediate control of HIV infection in both an independent and synergistic manner (Gaudieri et al. 2005; Martin et al. 2002, 2007). The recent characterisation of KIR haplotypes in MCM (Bimber et al. 2008) will facilitate detailed study of this gene family in relation to control of SIV infection and the restricted MHC diversity of MCM and availability of selectively bred MHC-matched animals (Mee et al. 2009) will facilitate study of KIR–MHC interactions.
In summary, we have performed the largest study to date of the effects of Mhc haplotypes on the outcome of SIV infection in a cohort of Mauritian cynomolgus macaques, and shown that haplotype H6 is associated with lower chronic phase viral loads. In the context of HIV vaccine development, our findings represent an important step towards the goal of defining the effects of host genetics on viral containment in a major animal model of human disease, and provide a foundation for more extensive studies of cellular immunity in cynomolgus macaques.
References
Almond N, Stott J (1999) Live attenuated SIV—a model of a vaccine for AIDS. Immunol Lett 66:167–170. doi:10.1016/S0165-2478(98) 00153-9
Berry N, Stebbings R, Brown S, Christian P, Thorstensson R, Ahmed RK, Davis L, Ferguson D, D'Arcy N, Elsley W, Hull R, Lines J, Wade-Evans A, Stott J, Almond N (2007) Immunological responses and viral modulatory effects of vaccination with recombinant modified vaccinia virus Ankara (rMVA) expressing structural and regulatory transgenes of simian immunodeficiency virus (SIVmac32H/J5M). J Med Primatol 36:80–94. doi:10.1111/j.1600-0684.2007.00216.x
Berry N, Stebbings R, Ferguson D, Ham C, Alden J, Brown S, Jenkins A, Lines J, Duffy L, Davis L, Elsley W, Page M, Hull R, Stott J, Almond N (2008) Resistance to superinfection by a vigorously replicating, uncloned stock of simian immunodeficiency virus (SIVmac251) stimulates replication of a live attenuated virus vaccine (SIVmacC8). J Gen Virol 89:2240–2251. doi:10.1099/vir.0.2008/001693-0
Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, Woodberry T, Sango K, Hewitt HS, Henry L, Linde CH, Chisholm JV 3rd, Zaman TM, Pae E, Mallal S, Walker BD, Sette A, Korber BT, Heckerman D, Brander C (2006) Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol 176:4094–4101
Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O'Connor DH (2008) Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol 181:6301–6308
Borsetti A, Baroncelli S, Maggiorella MT, Bellino S, Moretti S, Sernicola L, Belli R, Ridolfi B, Farcomeni S, Negri DR, Cafaro A, Ensoli B, Titti F (2008) Viral outcome of simian-human immunodeficiency virus SHIV-89.6P adapted to cynomolgus monkeys. Arch Virol 153:463–472. doi:10.1007/s00705-007-0009-2
Brandin E, Thorstensson R, Bonhoeffer S, Albert J (2006) Rapid viral decay in simian immunodeficiency virus-infected macaques receiving quadruple antiretroviral therapy. J Virol 80:9861–9864. doi:10.1128/JVI.00394-06
Dittmer U, Petry H, Stahl-Hennig C, Nisslein T, Spring M, Luke W, Bodemer W, Kaup FJ, Hunsmann G (1996) T cell apoptosis in human immunodeficiency virus type 2- and simian immunodeficiency virus-infected macaques. J Gen Virol 77:2433–2436. doi:10.1099/0022-1317-77-10-2433
Feichtinger H, Putkonen P, Parravicini C, Li SL, Kaaya EE, Bottiger D, Biberfeld G, Biberfeld P (1990) Malignant lymphomas in cynomolgus monkeys infected with simian immunodeficiency virus. Am J Pathol 137:1311–1315
Florese RH, Wiseman RW, Venzon D, Karl JA, Demberg T, Larsen K, Flanary L, Kalyanaraman VS, Pal R, Titti F, Patterson LJ, Heath MJ, O'Connor DH, Cafaro A, Ensoli B, Robert-Guroff M (2008) Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: influence of Mauritian MHC haplotypes on susceptibility/resistance to SHIV(89.6P) infection. Vaccine 26:3312–3321. doi:10.1016/j.vaccine.2008.03.100
Gaudieri S, DeSantis D, McKinnon E, Moore C, Nolan D, Witt CS, Mallal SA, Christiansen FT (2005) Killer immunoglobulin-like receptors and HLA act both independently and synergistically to modify HIV disease progression. Genes Immun 6:683–690
Gillespie GM, Kaul R, Dong T, Yang HB, Rostron T, Bwayo JJ, Kiama P, Peto T, Plummer FA, McMichael AJ, Rowland-Jones SL (2002) Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 16:961–972. doi:10.1097/00002030-200205030-00002
Giraldo-Vela JP, Rudersdorf R, Chung C, Qi Y, Wallace LT, Bimber B, Borchardt GJ, Fisk DL, Glidden CE, Loffredo JT, Piaskowski SM, Furlott JR, Morales-Martinez JP, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI (2008) The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J Virol 82:859–870. doi:10.1128/JVI.01816-07
Gotch FM, Hovell R, Delchambre M, Silvera P, McMichael AJ (1991) Cytotoxic T-cell response to simian immunodeficiency virus by cynomolgus macaque monkeys immunized with recombinant vaccinia virus. AIDS 5:317–320. doi:10.1097/00002030-199103000-00012
Habis A, Baskin GB, Murphey-Corb M, Levy LS (1999) Simian AIDS-associated lymphoma in rhesus and cynomolgus monkeys recapitulates the primary pathobiological features of AIDS-associated non-Hodgkin's lymphoma. AIDS Res Hum Retroviruses 15:1389–1398. doi:10.1089/088922299310098
Hardie RA, Knight E, Bruneau B, Semeniuk C, Gill K, Nagelkerke N, Kimani J, Wachihi C, Ngugi E, Luo M, Plummer FA (2008a) A common human leucocyte antigen-DP genotype is associated with resistance to HIV-1 infection in Kenyan sex workers. AIDS 22:2038–2042. doi:10.1097/QAD.0b013e328311d1a0
Hardie RA, Luo M, Bruneau B, Knight E, Nagelkerke NJ, Kimani J, Wachihi C, Ngugi EN, Plummer FA (2008b) Human leukocyte antigen-DQ alleles and haplotypes and their associations with resistance and susceptibility to HIV-1 infection. AIDS 22:807–816. doi:10.1097/QAD.0b013e3282f51b71
Jorajuria S, Clayette P, Dereuddre-Bosquet N, Larghero J, Thiebot H, Neildez O, Vaslin B, Le Grand R, Dormont D (2000) Evaluation of the effect of early and massive tritherapy on the expression of cellular factors potentially implicated in antiretroviral therapy resistance. Pathol Biol (Paris) 48:490–494
Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL (1996) Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med 2:405–411. doi:10.1038/nm0496-405
Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ (2004) Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775. doi:10.1038/nature03113
Lacap PA, Huntington JD, Luo M, Nagelkerke NJ, Bielawny T, Kimani J, Wachihi C, Ngugi EN, Plummer FA (2008) Associations of human leukocyte antigen DRB with resistance or susceptibility to HIV-1 infection in the Pumwani Sex Worker Cohort. AIDS 22:1029–1038. doi:10.1097/QAD.0b013e3282ffb3db
Le Grand R, Clayette P, Noack O, Vaslin B, Theodoro F, Michel G, Roques P, Dormont D (1994) An animal model for antilentiviral therapy: effect of zidovudine on viral load during acute infection after exposure of macaques to simian immunodeficiency virus. AIDS Res Hum Retroviruses 10:1279–1287
Loffredo JT, Friedrich TC, Leon EJ, Stephany JJ, Rodrigues DS, Spencer SP, Bean AT, Beal DR, Burwitz BJ, Rudersdorf RA, Wallace LT, Piaskowski SM, May GE, Sidney J, Gostick E, Wilson NA, Price DA, Kallas EG, Piontkivska H, Hughes AL, Sette A, Watkins DI (2007a) CD8 T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS One 2:e1152. doi:10.1371/journal.pone.0001152
Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI (2007b) Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol 81:8827–8832. doi:10.1128/JVI.00895-07
Maggiorella MT, Monardo F, Koanga-Mogtomo ML, Cioe L, Sernicola L, Corrias F, Baroni CD, Verani P, Titti F (1998) Detection of infectious simian immunodeficiency virus in B- and T-cell lymphomas of experimentally infected macaques. Blood 91:3103–3111
Maggiorella MT, Sernicola L, Crostarosa F, Belli R, Pavone-Cossut MR, Macchia I, Farcomeni S, Tenner-Racz K, Racz P, Ensoli B, Titti F (2007) Multiprotein genetic vaccine in the SIV-Macaca animal model: a promising approach to generate sterilizing immunity to HIV infection. J Med Primatol 36:180–194. doi:10.1111/j.1600-0684.2007.00236.x
Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M (2002) Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31:429–434
Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien S, Walker J, Parham BD, Deeks P, Mc SG, Vicar DW, Carrington M (2007) Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740. doi:10.1038/ng2035
McNeil AJ, Yap PL, Gore SM, Brettle RP, McColl M, Wyld R, Davidson S, Weightman R, Richardson AM, Robertson JR (1996) Association of HLA types A1-B8-DR3 and B27 with rapid and slow progression of HIV disease. QJM 89:177–185
Mee ET, Murrell CK, Sauermann U, Wilkinson RC, Cutler K, North D, Heath A, Ladhani K, Almond N, Rose NJ (2008) The Mhc class II DRB genotype of Macaca fascicularis does not influence infection by simian retrovirus type 2. Tissue Antigens 72:369–378. doi:10.1111/j.1399-0039.2008.01114.x
Mee ET, Badhan A, Karl JA, Wiseman RW, Cutler K, Knapp LA, Almond N, O'Connor DH, Rose NJ (2009) MHC haplotype frequencies in a UK breeding colony of Mauritian cynomolgus macaques mirror those found in a distinct population from the same geographic origin. J Med Primatol 38:1–14. doi:10.1111/j.1600-0684.2008.00299.x
Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M (2000) HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A 97:2709–2714. doi:10.1073/pnas.050567397
Montgomery MM, Dean AF, Taffs F, Stott EJ, Lantos PL, Luthert PJ (1999) Progressive dendritic pathology in cynomolgus macaques infected with simian immunodeficiency virus. Neuropathol Appl Neurobiol 25:11–19. doi:10.1046/j.1365-2990.1999.00163.x
Mothe BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, Allison DB, Watkins DI (2003) Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol 77:2736–2740. doi:10.1128/JVI.77.4.2736-2740.2003
Muhl T, Krawczak M, Ten Haaft P, Hunsmann G, Sauermann U (2002) MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J Immunol 169:3438–3446
Ndung'u T, Gaseitsiwe S, Sepako E, Doualla-Bell F, Peter T, Kim S, Thior I, Novitsky VA, Essex M (2005) Major histocompatibility complex class II (HLA-DRB and -DQB) allele frequencies in Botswana: association with human immunodeficiency virus type 1 infection. Clin Diagn Lab Immunol 12:1020–1028. doi:10.1128/CDLI.12.9.1020-1028.2005
Negri DR, Baroncelli S, Catone S, Comini A, Michelini Z, Maggiorella MT, Sernicola L, Crostarosa F, Belli R, Mancini MG, Farcomeni S, Fagrouch Z, Ciccozzi M, Boros S, Liljestrom P, Norley S, Heeney J, Titti F (2004) Protective efficacy of a multicomponent vector vaccine in cynomolgus monkeys after intrarectal simian immunodeficiency virus challenge. J Gen Virol 85:1191–1201. doi:10.1099/vir.0.79794-0
Neildez O, Le Grand R, Caufour P, Vaslin B, Cheret A, Matheux F, Theodoro F, Roques P, Dormont D (1998) Selective quasispecies transmission after systemic or mucosal exposure of macaques to simian immunodeficiency virus. Virology 243:12–20. doi:10.1006/viro.1997.9026
Nelson GW, Kaslow R, Mann DL (1997) Frequency of HLA allele-specific peptide motifs in HIV-1 proteins correlates with the allele's association with relative rates of disease progression after HIV-1 infection. Proc Natl Acad Sci U S A 94:9802–9807. doi:10.1073/pnas.94.18.9802
Nilsson C, Makitalo B, Berglund P, Bex F, Liljestrom P, Sutter G, Erfle V, ten Haaft P, Heeney J, Biberfeld G, Thorstensson R (2001) Enhanced simian immunodeficiency virus-specific immune responses in macaques induced by priming with recombinant Semliki Forest virus and boosting with modified vaccinia virus Ankara. Vaccine 19:3526–3536. doi:10.1016/S0264-410X(01) 00034-2
O'Connor DH, Mothe BR, Weinfurter JT, Fuenger S, Rehrauer WM, **g P, Rudersdorf RR, Liebl ME, Krebs K, Vasquez J, Dodds E, Loffredo J, Martin S, McDermott AB, Allen TM, Wang C, Doxiadis GG, Montefiori DC, Hughes A, Burton DR, Allison DB, Wolinsky SM, Bontrop R, Picker LJ, Watkins DI (2003) Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J Virol 77:9029–9040. doi:10.1128/JVI.77.16.9029-9040.2003
Putkonen P, Warstedt K, Thorstensson R, Benthin R, Albert J, Lundgren B, Oberg B, Norrby E, Biberfeld G (1989) Experimental infection of cynomolgus monkeys (Macaca fascicularis) with simian immunodeficiency virus (SIVsm). J Acquir Immune Defic Syndr 2:359–365
Putkonen P, Quesada-Rolander M, Leandersson AC, Schwartz S, Thorstensson R, Okuda K, Wahren B, Hinkula J (1998) Immune responses but no protection against SHIV by gene-gun delivery of HIV-1 DNA followed by recombinant subunit protein boosts. Virology 250:293–301. doi:10.1006/viro.1998.9379
Stebbings RJ, Almond NM, Stott EJ, Berry N, Wade-Evans AM, Hull R, Lines J, Silvera P, Sangster R, Corcoran T, Rose J, Walker KB (2002) Mechanisms of protection induced by attenuated simian immunodeficiency virus. Virology 296:338–353. doi:10.1006/viro.2002.1379
Stebbings R, Berry N, Stott J, Hull R, Walker B, Lines J, Elsley W, Brown S, Wade-Evans A, Davis G, Cowie J, Sethi M, Almond N (2004) Vaccination with live attenuated simian immunodeficiency virus for 21 days protects against superinfection. Virology 330:249–260. doi:10.1016/j.virol.2004.09.026
ten Haaft P, Verstrepen B, Uberla K, Rosenwirth B, Heeney J (1998) A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J Virol 72:10281–10285
ten Haaft P, Almond N, Biberfeld G, Cafaro A, Cranage M, Ensoli B, Hunsmann G, Polyanskaya N, Stahl-Hennig C, Thortensson R, Titti F, Heeney J (2001) Comparison of early plasma RNA loads in different macaque species and the impact of different routes of exposure on SIV/SHIV infection. J Med Primatol 30:207–214. doi:10.1034/j.1600-0684.2001.d01-54.x
Titti F, Sernicola L, Geraci A, Panzini G, Di Fabio S, Belli R, Monardo F, Borsetti A, Maggiorella MT, Koanga-Mogtomo M, Corrias F, Zamarchi R, Amadori A, Chieco-Bianchi L, Verani P (1997) Live attenuated simian immunodeficiency virus prevents super-infection by cloned SIVmac251 in cynomolgus monkeys. J Gen Virol 78(Pt 10):2529–2539
Tsai CC, Emau P, Follis KE, Beck TW, Benveniste RE, Bischofberger N, Lifson JD, Morton WR (1998) Effectiveness of postinoculation (R)-9-(2-phosphonylmethoxypropyl) adenine treatment for prevention of persistent simian immunodeficiency virus SIVmne infection depends critically on timing of initiation and duration of treatment. J Virol 72:4265–4273
Wade-Evans AM, Stott J, Hanke T, Stebbings R, Berry N, Lines J, Sangster R, Silvera P, Walker B, MacManus S, Davis G, Cowie J, Arnold C, Hull R, Almond N (2001) Specific proliferative T cell responses and antibodies elicited by vaccination with simian immunodeficiency virus Nef do not confer protection against virus challenge. AIDS Res Hum Retroviruses 17:1517–1526. doi:10.1089/08892220152644223
Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O'Connor SL, O'Connor DH (2007) Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol 81:349–361. doi:10.1128/JVI.01841-06
Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI (2006) The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol 80:5074–5077. doi:10.1128/JVI.80.10.5074-5077.2006
Zhang ZQ, Fu TM, Casimiro DR, Davies ME, Liang X, Schleif WA, Handt L, Tussey L, Chen M, Tang A, Wilson KA, Trigona WL, Freed DC, Tan CY, Horton M, Emini EA, Shiver JW (2002) Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J Virol 76:12845–12854. doi:10.1128/JVI.76.24.12845-12854.2002
Acknowledgements
We are grateful to the veterinary and support staff, and to Alan Heath for assistance with statistical analysis. This work was funded by the UK Department of Health; ENVEP, an EU Framework Programme 5 grant QLK2-CT-1999-00871; EUPRIM-NET, an EU Framework Programme 6 grant and Medical Research Council grant G9025730.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1
Treatment regimens and infection outcome of animals included in study (DOC 39 kb)
Rights and permissions
About this article
Cite this article
Mee, E.T., Berry, N., Ham, C. et al. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics 61, 327–339 (2009). https://doi.org/10.1007/s00251-009-0369-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-009-0369-8