Abstract
Purpose
Early accurate diagnosis of infection ± organ dysfunction (sepsis) remains a major challenge in clinical practice. Utilizing effective biomarkers to identify infection and impending organ dysfunction before the onset of clinical signs and symptoms would enable earlier investigation and intervention. To our knowledge, no prior study has specifically examined the possibility of pre-symptomatic detection of sepsis.
Methods
Blood samples and clinical/laboratory data were collected daily from 4385 patients undergoing elective surgery. An adjudication panel identified 154 patients with definite postoperative infection, of whom 98 developed sepsis. Transcriptomic profiling and subsequent RT-qPCR were undertaken on sequential blood samples taken postoperatively from these patients in the three days prior to the onset of symptoms. Comparison was made against postoperative day-, age-, sex- and procedure- matched patients who had an uncomplicated recovery (n =151) or postoperative inflammation without infection (n =148).
Results
Specific gene signatures optimized to predict infection or sepsis in the three days prior to clinical presentation were identified in initial discovery cohorts. Subsequent classification using machine learning with cross-validation with separate patient cohorts and their matched controls gave high Area Under the Receiver Operator Curve (AUC) values. These allowed discrimination of infection from uncomplicated recovery (AUC 0.871), infectious from non-infectious systemic inflammation (0.897), sepsis from other postoperative presentations (0.843), and sepsis from uncomplicated infection (0.703).
Conclusion
Host biomarker signatures may be able to identify postoperative infection or sepsis up to three days in advance of clinical recognition. If validated in future studies, these signatures offer potential diagnostic utility for postoperative management of deteriorating or high-risk surgical patients and, potentially, other patient populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Early transcriptomic changes offer potential diagnostic utility for the management of patients at risk of develo** subsequent postoperative infection ± sepsis. The limited number of genes identified facilitates the development of a point-of-care rapid diagnostic. |
Introduction
Sepsis, the dysregulated host response to infection leading to life-threatening organ dysfunction [1], is a substantial global cause of mortality and morbidity [2]. Early, accurate diagnosis of infection and organ dysfunction remains problematic, as reflected by multiple interventional trials over three decades failing to yield outcome improvements [3]. These failures relate in part to the uncertainty that infection was actually present [4, 5], but also to belated intervention once the patient is in established multi-organ failure with the therapeutic window of opportunity having closed [3, 6]. Success with immunomodulatory and other therapeutic strategies is likely predicated on use early in the sepsis course or even pre-emptively, as frequently achieved in preclinical models [6]. On the other hand, inappropriate use of antibiotics contributes to antimicrobial resistance [7] and may distract clinicians from diagnosing a non-infective condition [4, 5].
Considerable efforts are being expended to develop rapid, even point-of-care, biomarkers for infection and sepsis with high sensitivity and specificity [8]. Pathogen-focused techniques include molecular identification of the pathogen or unique components such as prokaryotic DNA. The dysregulated host response is being interrogated using -omic approaches, single- or multiplex protein assays or flow cytometry [8, 9]. However, host response studies to date often lack comparator cohorts of patients with non-infective causes of critical illness or microbiological confirmation that infection is indeed present. Furthermore, sampling is usually begun after the patient presents with suspected infection or sepsis. An ideal patient cohort would allow accurate pre-symptomatic identification of patients who proceed to develop an infection, in particular those progressing to sepsis.
Our primary study aim was to develop a tool to facilitate presymptomatic identification of patients develo** an infection, and we hypothesised this could be achieved using transcriptional changes in small gene sets. As a secondary objective, we sought to determine whether we could pre-identify those infected patients who would develop new-onset organ dysfunction (sepsis). To this end, we conducted a large prospective, multi-centre study in patients undergoing elective major surgery, with daily blood sampling and data recording commencing preoperatively and continuing up to a week after. A clinical adjudication panel independently examined clinical and laboratory data to identify patients with definite infection ± sepsis. Samples from these patients enabled comparison of microarray and RT-qPCR data against cohorts of postoperative day-, age-, sex- and procedure-matched patients with either non-infective systemic inflammation or an uncomplicated postoperative course.
Materials and methods
Study design
Elective surgery patients were prospectively recruited at eight hospitals (seven UK, one German) between November 2007 and February 2017. Ethical approval was granted through the Southampton and South-West Hampshire Multicentre Research Ethics Committee (Reference 06/Q1702/152). The protocol achieved US Federal Wide Assurance Independent Review Board status (IRB00001756). Patients were enrolled if they gave informed consent, were aged between 18-80 years, and due to undergo elective major surgery that would likely enhance the development of postoperative infection (Supplement S1).
Data and blood sampling collection
Demographic data were collected at enrolment with baseline blood sample collection taken at 1-7 days pre-operatively. After surgery, relevant clinical, laboratory and imaging data and blood samples were collected daily on all patients until seven days, hospital discharge (if sooner), or a diagnosis of infection or sepsis by the treating clinician. Two 4 ml aliquots of blood were collected daily into sterile EDTA vacutainers and then immediately transferred into RNAse-free vials containing 10.5 ml RNAlater® (ThermoFisher, Waltham, MA, USA) to preserve the transcriptome.
Patient selection process
A detailed description is provided in Supplement S2. Briefly, the initial diagnosis of sepsis was based on the treating clinician’s interpretation of clinical and laboratory markers using the then-extant ‘Sepsis-2’ definition of sepsis [10]. This described ‘sepsis’ as suspected or confirmed infection with two or more systemic inflammatory response syndrome (SIRS) criteria, and ‘severe sepsis’ as sepsis plus poorly-characterized new-onset organ dysfunction. The new sepsis definition (Sepsis-3), published in February 2016 redefined ‘sepsis’ as infection plus new organ dysfunction identified by a ≥2 point rise in the Sequential Organ Failure Assessment (SOFA) score [1]. To align with modern nomenclature, subsequent analyses and descriptors apply the new definition.
Since clinical opinion can vary when diagnosing infection or sepsis [11], a Clinical Advisory Panel adjudicated cases and enabled confident identification of patients with postoperative infection (Supplement S2). Patients with definite infection were allocated to either an uncomplicated infection subgroup or to a sepsis subgroup if new organ dysfunction developed following surgery with a rise in SOFA score ≥2 points above baseline [1]. Postoperative day-, age-, sex- and procedure-matched cohorts of non-infected patients making an uncomplicated postoperative recovery (SIRS-), or develo** a systemic inflammatory response (SIRS+), were selected from the remaining pool of patients recruited into the study. Blood samples from these three groups, totalling 453 patients, were taken forward for transcriptomic and subsequent reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analyses.
Microarray and RT-qPCR analysis
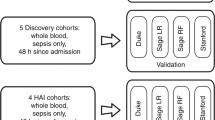
Infected patients were split into Discovery (Gene Feature Selection) (n= 63) and Training and Validation (Classification) (n=91) cohorts (Fig. 1).
Study schema with Discovery (Gene Feature Selection) and Training/Validation (Classification) phases indicated. RT-qPCR Reverse Transcription quantitative polymerase chain reaction, SIRS systemic inflammatory response syndrome, Inf+ infected patients, Inf- non-infected patients; Inf+ OD- uncomplicated infection patients
Discovery (Gene Feature Selection) Cohort
Biomarker discovery studies were undertaken exclusively in this 63-patient cohort at a pre-determined interim analysis of the study. A detailed transcriptomic analysis was initially undertaken using globin-reduced RNA (GlobinClear Human, ThermoFisher) from the blood samples of 58 patients collected during the three days prior to presentation of definite infection, and from 55 matched non-infected (SIRS-) control patients (Fig. 1). Further details of the microarray analysis using Human HT-12v4 bead arrays (Illumina, San Diego, CA, USA) are shown in Supplement S3. Differentially expressed genes (DEGs) based on microarray data were identified by applying a linear model fit for each gene [12], a p-value cut-off of 0.05 and a fold change of at least 1.2. Data obtained from non-infected study patients were used as reference. Gene expression of the top 80 identified DEGs was verified using multiplex RT-qPCR (Fluidigm, San Francisco, CA, USA) and TaqMan Gene Expression Assays (Applied Biosystems, Carlsbad, CA, USA).
The RT-qPCR data were then further used for down selecting the 25 most promising biomarkers. In these experiments, analysis was possible for 63 infection and 62 SIRS- patients. In addition, RT-qPCR data were obtained from 58 SIRS+ matched patients (Fig. 1; Table 1). Within the 63 infected patients, 37 develo** sepsis were compared to 26 with uncomplicated infection (Fig. 1; Table 1). Gene ontology enrichment was performed using the R package cluster profiler (v3.18.0) and visualized using the R package GoPlot (v1.0.2).
Down-selection of genes based on RT-qPCR data was made using a two-step feature selection process to improve the performance of the predictive classification models used to discern patient groups (described fully in Supplement S4). Briefly, the Boruta algorithm [13], a wrapper method based on random forest [14] was used for the selection of relevant features in the dataset. This step was followed by backward elimination to determine features with the most discriminatory power for a particular classification. Training of these models relevant for feature selection was done by a 5-fold cross-validation repeated 25 times. Mean performances were tracked for Area Under the Receiver Operator Curve (AUC), PPV and NPV over the 25 repetitions. This procedure was repeated to assess the ability of the identified biomarkers to discriminate between infected patients and either controls (SIRS-) or non-infected but inflamed (SIRS+) patients and, as a secondary objective, to discriminate between infected patients either develo** organ dysfunction (sepsis) or not (Fig. 1).
Training and validation of final classification models
Of all 80 DEG biomarker candidates, 25 were identified as part of at least one classification model based on an orthogonal RT-qPCR approach that could discriminate between the patient classes. These 25 RT-qPCR transcripts were taken forward to an independent patient cohort (Classification Cohort) to both learn a predictive classifier model and to assess performance.
RT-qPCR analysis of gene expression was undertaken on samples from 91 patients with infection, 89 matched SIRS- comparator patients, and 90 similarly matched non-infected SIRS+ patients, respectively (Fig. 1). Further comparison was made within the infection cohort, comparing patients who did (sepsis, n=61) or did not develop organ dysfunction (n=30). A final comparison was made between patients with sepsis (n=61) and all other matched patients with non-sepsis outcomes (n=209) within our validation cohorts (Fig. 1).
For assessment of signature performance, a similar random forest-based approach was used. Models were created (‘training phase’) and then validated (‘validation phase’). We divided our data in a random 80:20 split for training and validation, respectively. In the training phase models were trained and cross-validated on the 80% subset using 5-fold cross-validation. This process was repeated 25 times using R package caret (v6.0-90, ‘train’ function) to prevent lucky or unlucky 5-fold cross-validation. In the validation phase, the trained models were tested on the remaining 20% of the data that were not used for cross-validation in the training phase. This process was repeated 10 times with 10 different randomly selected subsets, again to counteract any lucky or unlucky data splits. For each comparison, 10 models (including 10 different test sets) were created and the mean AUC, sensitivity and specificity reported.
Statistical analysis
Prediction of a robust sample size followed the method described by Figueroa et al [15] and is described in detail in Supplement S4. Briefly, random forest-based classifier performance was assessed with an increasing number of simulated patients in each cohort to identify when a performance plateau occurred.
All qRT-PCR data were analyzed with nonparametric Wilcoxon tests unless otherwise stated. All data and statistical analyses were done using R (v.3.6.3).
Results
Patient recruitment and subgroup identification
Clinical data and blood samples were collected from 4385 high-risk elective surgery patients. An overall flow chart is shown in Supplement S5. The clinical panel adjudication identified a total of 154 patients with high certainty of infection, of whom 98 developed sepsis (Fig. 1). These were compared against samples taken from the postoperative day-, age-, sex- and operation-matched patients without infection, of whom 148 developed a systemic inflammatory response (SIRS+) and 151 (SIRS-) did not. Each patient group was split into Discovery (Gene Feature selection) and Training/Validation (Classification) cohorts. Demographic and clinical data are shown in Table 1. Microbiological confirmation of infection was achieved in 81 septic and 45 uncomplicated infection patients (Supplement S6). A broad range of Gram negative, Gram positive and fungal organisms were isolated. Strong clinical evidence of infection was used for the remaining 15 and 11 patients, respectively. Of the 98 septic patients, 80 patients required at least one organ support (60 mechanical ventilation, 55 vasopressors, 8 renal replacement therapy) Nine patients ultimately died of sepsis while two died for unrelated reasons. Approximately 60% of the non-infected patients received no postoperative antibiotic (Supplement S7). The remaining patients received one or more doses of antibiotic, 79% commencing on Postoperative Day 1 and likely to represent ongoing prophylactic medication.
Discovery (Gene Feature selection) cohort
Microarray-based biomarker discovery
Initial biomarker discovery was performed on blood samples taken from 58 infected patients over the three days preceding the clinical presentation of postoperative infection and underwent transcriptomic profiling. Comparison was made against 55 postoperative day-, age-, sex- and procedure-matched patients with an uncomplicated, non-infected, non-inflamed (SIRS-) course (Fig. 1, Table 1).
Overall, 2594 differentially expressed genes (DEGs) between infection and control were identified in the three days pre-infection diagnosis. Of the top 1500 DEGs with the highest fold change (Fig. 2A, Supplement S8), 863 (57.5%) DEGs were upregulated and 637 (42.5%) downregulated. Functional enrichment analysis yielded immune relevant pathways involving primarily neutrophils and T cells (Fig. 2B, Supplement S8).
Gene expression analysis for infection versus healthy postoperative controls. A Expression profile of 1500 differentially expressed genes with the highest absolute fold change. The top ten up- and downregulated genes are indicated separately. B Top enriched Gene Ontology categories based on 1500 differentially expressed genes with the highest fold change as shown in Fig. 2A). Node sizes indicate a number of genes per shown category. Fold change is indicated for each displayed gene node. C RT-qPCR values for 8 genes originating from classifying infection versus no infection based on microarray-based expression data. ΔCq value indicates PCR cycle quantifications according to ΔCq = Cq(reference) - Cq(target gene). Wilcoxon test derived significance level is indicated (***p≤0.001, ns: not significant). Error bars show standard error of the mean. N(samples of infected patients) = 139, N(samples of control patients) = 144
A random forest-based algorithm identified the best-performing DEGs derived for all three days prior to infection/sepsis diagnosis and their respective non-infected controls. Initial analysis indicated that different sets of eight genes could identify postoperative infection with high AUCs.
RT-qPCR verification and down-selection of microarray-based biomarkers
Collectively, differential expression analysis and concurrent random forest-based classifiers with equally high-performance metrics proposed 80 best performing genes from the microarray data. These were down-selected for confirmation by RT-qPCR analysis using the same 58 infected patient cohort samples analysed for biomarker discovery plus samples from an additional five patients, with comparisons being made against 62 non-infected SIRS- patients. A representative 8-gene-set showed highly significant differential expression between the two patient groups, exemplifying the compatibility between microarray- and RT-qPCR-based gene expression (FDR-corrected Wilcoxon test, p<0.001, Fig. 2C). To utilise all patient sub-groups with different postoperative outcomes identified in our study, we subsequently explored whether subsets of our 80-gene set would suffice to differentiate the 63 infected patients from 58 SIRS+ patients, as well as to differentiate within the infection cohort, i.e. with (n=37) or without (n = 26) organ dysfunction (Fig. 1; Table 1). Again using a random forest-based classification approach in conjunction with Boruta to select the most important gene transcripts (cf Methods) yielded convincing model performances throughout all classification attempts. Specifically, a 7 gene set (B4GALT5, AFF1, LDLR, ATXN7L3, LARP4B, SLC36A1, TRPM2, AUC >0.85, PPV >0.8) for infection versus SIRS- models, a 12 gene set (ATXN1, SLC41A3, MED13L, STOM, B4GALT5, MIDN, HVCN1, LDLR, CFLAR, SPATA13, EIF4G3, METTL7B, AUC >0.9, PPV >0.8) for infection versus SIRS+ models, and an eight gene set (DOK3, ICAM2, IL1R1, LGALS2, LSG1, RPL13A, RPS13, SGSH, AUC >0.75 (PPV >0.7) for sepsis vs. uncomplicated infection were sufficient to classify postoperative outcomes. These 25 genes were further selected for building predictive models in an independent and substantially larger patient cohort.
Training/Validation (Classification) Cohort
Building machine learning predictive models in independent patient cohorts
In separate cohorts of patients, we evaluated the classification performances of the 7, 12 and 8 gene signatures whose performance had been verified to discern: (i) infection (n=91) from non-infected SIRS- (n=89) patients, (ii) infection from non-infected SIRS+ (n=90) patients, and (iii) 61 sepsis and 30 uncomplicated infection patients, respectively, (Fig. 1; Table 1).
To prevent models from overfitting, classification training including nested cross-validation was performed 10 times on a randomly selected 80% of the data. In each iteration, the models were validated on 20% of randomly selected sample subsets that were never used for training (kept outside of cross-validation, cf. Methods). The mean performance of the gene signatures across these 10 runs remained high following a random forest-based classification. For postoperative infection versus SIRS- controls, the 7 gene signature achieved an AUC value of 0.871 (Fig. 3). The 12 gene signature for differentiation of infection from non-infected SIRS+ patients achieved an AUC value of 0.897. Differentiation of sepsis from uncomplicated infection patients resulted in an AUC value of 0.703. Finally, differentiation of sepsis from all other clinical presentations using all 25 transcripts achieved an AUC of 0.843. Sensitivity was high across all comparisons (0.785-0.942) whereas specificity was high for infected compared to non-infected SIRS+ (0.838) and SIRS- (0.776) patients, but poor for sepsis vs uncomplicated infection (0.217). Supplement S9 shows the Classification performance for different thresholds optimized for sensitivity or specificity and various assumed prevalence of outcomes. This indicates, for example, how current practice specificity for correct antibiotic selection (Supplement 7) can be enhanced by the use of a specificity-optimized gene panel.
RT-qPCR based classification performance based on random forest using all available patient samples and across all days prior to infection diagnosis. ROC curve and statistical metrics for (i) infection versus non-inflamed postoperative controls (SIRS-) [blue], infection versus non-infected and inflamed (SIRS+) [red], sepsis versus infection without organ dysfunction (OD-) [mauve] and sepsis versus all others [green]. The table reports mean statistics for classification models based on different numbers of transcripts. N number, AUC area under the curve, PPV positive predictive value, NPV negative predictive value, Sens sensitivity, Spec specficity
Discussion
This study demonstrates that patients undergoing elective major surgery who develop a postoperative infection with or without organ dysfunction (sepsis), can be reliably identified and differentiated from non-infected patients by using host transcriptomics up to three days before clinical diagnosis.
Accurate, early diagnosis of infection and sepsis represents a holy grail for improving patient care and outcomes and avoiding unnecessary antimicrobial use. Transcriptomic studies of patients with uncomplicated infection and/or sepsis have focused on patients with established clinical features requiring hospitalization or admission to critical care [17,18,19,20,21]. Such studies have added to our understanding of sepsis pathogenesis, demonstrating some prognostic capability [18, 22,23,24], and offering reasonable discrimination between infectious and non-infectious causes of systemic inflammation [18,19,20, 25]. Presymptomatic diagnosis would facilitate early investigation and treatment, and can be reasonably surmised to improve patient outcomes.
To our knowledge, no study has specifically studied patient samples taken before the clinical onset of infection. An elective surgical population represents an ideal patient group, albeit somewhat inefficient, as most have a postoperative course uncomplicated by infection. Apart from the underlying need for surgery, patients enrolled in our study were infection-free and stable. To have as clean a dataset as possible, samples were only used from the 3.5% of patients where a clinical adjudication panel identified postoperative infection with high confidence. Intermediate cases with more uncertainty surrounding diagnosis were excluded. We also drew from a large patient cohort from which postoperative day-, age-, sex- and operation-matched patients could be selected having either an uncomplicated course or develo** a postoperative systemic inflammation where the infection was confidently excluded and not treated.
Many studies examining sepsis biomarkers have relied on selection through knowledge-based approaches predicated on known biological functions and pathways [25] rather than targets whose functions have not been fully described [26]. We adopted a target selection strategy that relied on statistical rather than biological features [27]. A machine learning approach was used to select appropriate targets and classify patients based on host gene expression with comparison against a clear-cut clinical diagnosis to determine predictive accuracy. The initial phase of biomarker discovery identified 80 genes for which differences from other patient cohorts could be confirmed using an orthogonal approach (RT-qPCR). Re-classification based on RT-qPCR aimed at feature reduction for best gene selection in the same group of patients enabled further down-selection to 25 key biomarkers. Subsequent classification analysis revealed the potential for clinical application with an even more reduced set of biomarkers, allowing a rapid and affordable point-of-care test.
A machine learning algorithm approach was also used to select appropriate targets and classify patients based on host gene expression (uncorrected for baseline values) with a comparison made against a clear-cut clinical diagnosis to determine predictive accuracy. The success of this approach is evidenced by high values of AUC, sensitivity and specificity comparing patients develo** infection versus uncomplicated (SIRS-) controls, and the more diagnostically challenging group who develop postoperative (SIRS+) inflammation that is not driven by infection yet which shares many clinical features such as pyrexia, tachycardia, neutrophilia and elevated C-reactive protein levels. There was also reasonable discrimination (high sensitivity but poor specificity) in pre-emptively identifying infected patients who would proceed to organ dysfunction compared to those with uncomplicated infection.
The difference in our study approach is also exemplified by only six out of 80 genes identified in our classification models overlap** with genes of prior published gene signatures for sepsis and community-acquired pneumonia [16, 18, 20]. The data reported within our study indicate that changes in expression of host biomarkers are predictive of later complications related to infection. Any shortcomings in model performance are likely due to the complex relationship between individual genes, their transcripts, post-translational modifications and protein interactions, and complicated further by both therapeutic interventions and the dynamic nature of signaling within the host response that flux during the dysregulation process [27, 28].
In trying to answer the more challenging question of when an individual will develop infection or sepsis, we sampled at multiple timepoints. While a common gene set could detect changes over a 3-day period prior to clinical presentation, separate gene transcripts gave even stronger signals on a day-by-day basis (data not shown). This reflects observations made in trauma patients [ Cohort data are available in Table 1 and the Supplement. Microarray data are stored on Mendeley (https://doi.org/10.17632/rhc5s6zj88.3) and will be available on reasonable request. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M et al (2016) The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315:801–810. https://doi.org/10.1001/jama.2016.0287 Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR et al (2020) Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet 395:200–11. https://doi.org/10.1016/S0140-6736(19)32989-7 Vincent J-L, Sakr Y (2019) Clinical trial design for unmet clinical needs: a spotlight on sepsis. Expert Rev Clin Pharmacol 12:893–900. https://doi.org/10.1080/17512433.2019.1643235 Heffner AC, Horton JM, Marchick MR, Jones AE (2010) Etiology of illness in patients with severe sepsis admitted to the hospital from the emergency department. Clin Infect Dis 50:814–20. https://doi.org/10.1086/650580 Klein Klouwenberg PMC, Cremer OL, van Vught LA, Ong DS, Frencken JF, Schultz MJ et al (2015) Likelihood of infection in patients with presumed sepsis at the time of intensive care unit admission: a cohort study. Crit Care 19:319. https://doi.org/10.1186/s13054-015-1035-1 Perner A, Gordon AC, Angus DC, Lamontagne F, Machado F, Russell JA et al (2017) The intensive care medicine research agenda on septic shock. Intensive Care Med 43:1294–305. https://doi.org/10.1007/s00134-017-4821-1 World Health Organisation: Antimicrobial resistance, https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed 19 November 2021). Opal SM, Wittebole X (2020) Biomarkers of infection and sepsis. Crit Care Clin 36:11–22. https://doi.org/10.1016/j.ccc.2019.08.002 Kim M-H, Choi J-H (2020) An update on sepsis biomarkers. Infect Chemother 52:1–18. https://doi.org/10.3947/ic.2020.52.1.1 Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cohen J, Opal SM, Vincent JL, Ramsay G (2003) 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med 31:1250–6. https://doi.org/10.1097/01.CCM.0000050454.01978.3B Rhee C, Kadri SS, Danner RL, Suffredini AF, Massaro AF, Kitch BT et al (2016) Diagnosing sepsis is subjective and highly variable: a survey of intensivists using case vignettes. Crit Care 20:89. https://doi.org/10.1186/s13054-016-1266-9 Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47–e47. https://doi.org/10.1093/nar/gkv007 Kursa MB, Rudnicki WR (2010) Feature selection with the Boruta Package. J Stat Soft https://doi.org/10.18637/jss.v036.i11 Breiman L (2001) Random forest. Machine Learning 45:5–32 Figueroa RL, Zeng-Treitler Q, Kandula S, Ngo LH (2012) Predicting sample size required for classification performance. BMC Med Inform Decis Mak 12:8. https://doi.org/10.1186/1472-6947-12-8 Cazalis M-A, Lepape A, Venet F, Fragar F, Mougin B, Vallin H et al (2014) Early and dynamic changes in gene expression in septic shock patients: a genome-wide approach. Intensive Care Med Exp 2:20. https://doi.org/10.1186/s40635-014-0020-3 Langley RJ, Tsalik EL, Velkinburgh JC, Glickman SW, Rice BJ, Wang C et al (2013) An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. https://doi.org/10.1126/scitranslmed.3005893 Sweeney TE, Shidham A, Wong HR, Khatri P (2015) A comprehensive time-course–based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aaa5993 Miller RR, Lopansri BK, Burke JP, Levy M, Opal S, Rothman RE et al (2018) Validation of a host response assay, SeptiCyte LAB, for discriminating sepsis from systemic inflammatory response syndrome in the ICU. Am J Resp Crit Care Med 198:903–13. https://doi.org/10.1164/rccm.201712-2472OC Scicluna BP, Klein Klouwenberg PMC, van Vught LA, Wiewel MA, Ong DS, Zwinderman AH et al (2015) A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am J Resp Crit Care Med 192:826–35. https://doi.org/10.1164/rccm.201502-0355OC Burnham KL, Davenport EE, Radhakrishnan J, Humburg P, Gordon AC, Hutton P et al (2017) Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia. Am J Resp Crit Care Med 196:328–39. https://doi.org/10.1164/rccm.201608-1685OC Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM et al (2018) A community approach to mortality prediction in sepsis via gene expression analysis. Nature Commun 9:694. https://doi.org/10.1038/s41467-018-03078-2 Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL et al (2017) Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Resp Med 5:816–26. https://doi.org/10.1016/S2213-2600(17)30294-1 Pierrakos C, Vincent J-L (2010) Sepsis biomarkers: a review. Crit Care 14:R15. https://doi.org/10.1186/cc8872 Sutherland A, Thomas M, Brandon RA, Brandon RB, Lipman J, Tang B et al (2011) Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit Care 15:R149. https://doi.org/10.1186/cc10274 Kaplan JM, Wong HR (2011) Biomarker discovery and development in pediatric critical care medicine. Pediatr Crit Care Med 12:165–73. https://doi.org/10.1097/PCC.0b013e3181e28876 Maslove DM, Wong HR (2014) Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol Med 20:204–13. https://doi.org/10.1016/j.molmed.2014.01.006 Tang BMP, McLean AS, Dawes IW, Huang SJ, Lin RCY (2009) Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med 37:882–8. https://doi.org/10.1097/CCM.0b013e31819b52fd **ao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H et al (2011) A genomic storm in critically injured humans. J Exp Med 208:2581–90. https://doi.org/10.1084/jem.20111354 Calvano SE, **ao W, Richards DR, Felciano RM, Baker HV, Cho RJ et al (2005) (2005) A network-based analysis of systemic inflammation in humans. Nature 437:1032–7. https://doi.org/10.1038/nature03985 Kwan A, Hubank M, Rashid A, Klein N, Peters MJ (2013) Transcriptional instability during evolving sepsis may limit biomarker based risk stratification. PLoS One 8:e60501. https://doi.org/10.1371/journal.pone.0060501 This study was funded by: The US Defense Threat Reduction Agency, Pre-symptomatic map** of the host response that leads to sepsis (DTRA01-03-D-0001-0017) and “Correlating Pre-Symptomatic Biomarkers for Sepsis” (HDTRA1-12-D-0003-0011). These funded patient recruitment, sample collection and laboratory analysis of patient bloods (microarray and qRT-PCR). The UK Ministry of Defence and the UK Home Office. These funded patient recruitment, sample collection and laboratory analysis of patient bloods (RT-qPCR). This study was supported by researchers at the: National Institute for Health and Care Research University College London Hospitals Biomedical Research Centre; Benaroya Research Institute, Seattle; Queen Elizabeth Hospital, Birmingham; Bristol Royal Infirmary; University Hospital Frankfurt, Guy’s and St. Thomas’s NHS Foundation Trust; Heartlands hospital, Birmingham; Leeds General Infirmary and BioXpedia. We would also like to acknowledge the following individuals: Sample processing and curation: Wendy Butcher, Emma Keyser & Jaki Walsh (Dstl). Project Management: Ehsan Gazi, Amy Middleton, Holly Riley, Gary Walker & Polly Williams (Dstl). Patient Selection Analytics: Phillippa Spencer & Laura Pearsall (Dstl). Patient enrolment, sample and data collection: Katie Sweet (Bristol Royal Infirmary), Karen Williams, Anna Walker (Royal Liverpool Hospital), Arlo Whitehouse, Lauren Cooper (Queen Elizabeth Hospital, Birmingham), Jung Hyun Ryu (University College London Hospitals), Katie Lei (Guy’s & St Thomas’ Hospitals), Simone Landau, Carolin Widenbeck, Karin Pense (University Hospital Frankfurt, Goethe University). RT-qPCR @ BioXpedia: Hans-Christian Ingerslev. Finally, the authors would like to thank Julian Bion for his valuable insights over the course of this project. All authors contributed to the study’s conception and design. Material preparation, data collection and analysis were performed by RAL, HEJ, VHG, DB, JW, MT, TW, MO, AK, KZ, MK and DC. The first draft of the manuscript was written by RAL, MK and MS and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. RAL reports grants from the US Defense Threat Reduction Agency, UK Ministry of Defence and UK Home Office during the conduct of the study. He has a patent for Gene-based Methods and Associated Uses, Kits and System for Assessing Sepsis. GB2015002.5 Filed Sept. 2020 pending, a patent Gene-based Methods and Associated Uses, Kits and System for Assessing Sepsis. GB2015004.1 Filed Sept. 2020 pending, a patent Gene-based Methods and Associated Uses, Kits and System for Assessing Sepsis. GB2015007.4 Filed Sept. 2020 pending, a patent Sepsis Nucleic Acid Markers. GB1402293.3 Filed Jan 2014 issued, and a patent Protein-based Methods and Associated Uses, Kits and System for Assessing Sepsis. GB2009401.7 Filed June 2020, pending. He is a part-time advisor to PreSymptom Health Ltd and have licensed patents belonging to the UK Ministry of Defence. HJ - HEJ reports grants from the US Defense Threat Reduction Agency, grants from the UK Ministry of Defence, and grants from UK Home Office, during the conduct of the study. In addition, HEJ has a patent for Sepsis Nucleic Acid Markers. GB1402293.3 Filed Jan 2014 issued. MS - MS reports grants from the Defence Science and Technology Laboratory during the conduct of the study; grants from NewB, Apollo Therapeutics and UCL Technology Fund, others from Abbott, Amormed, Biomerieux, Biotest, Deltex Medical, Fresenius, Mindray, NewB, Pfizer, Radiometer, Roche Diagnostics, Safeguard Biosystems, Shionogi and Spiden outside of this project. He is an unpaid advisor to PreSymptom Health Ltd, Hemotune, deepUll, and Santersus. KZ reports grants from Defence Science and Technology Laboratory, during the conduct of the study; grants and other from EU-Horizon 2020 Projekt ENVISION, Horizon Europe 2021 Projekt COVend, DFG, BMBF, ECCPS, LOEWE, B. Braun Melsungen AG; Biotest AG; CSL Behring GmbH; Fresenius Kabi GmbH; Fresenius Medical Care; Haemonetics; Löwenstein Medical GmbH; med update GmbH; Vifor Pharma GmbH; Werfen. No other author reported any potential conflicts of interest. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Below is the link to the electronic supplementary material. Lukaszewski, R.A., Jones, H.E., Gersuk, V.H. et al. Presymptomatic diagnosis of postoperative infection and sepsis using gene expression signatures.
Intensive Care Med 48, 1133–1143 (2022). https://doi.org/10.1007/s00134-022-06769-z Received: Accepted: Published: Issue Date: DOI: https://doi.org/10.1007/s00134-022-06769-zData sharing
References
Acknowledgements
Author information
Authors and Affiliations
Contributions
Corresponding author
Ethics declarations
Conflicts of interest
Additional information
Publisher's Note
Supplementary Information
Rights and permissions
About this article
Cite this article
Keywords