Abstract
Background
N6-methyladenosine (m6A) RNA modification regulators play an important role in many human diseases, and its abnormal expression can lead to the occurrence and development of diseases. However, their significance in pulpitis remains largely unknown. Here, we sought to identify and validate the m6A RNA regulatory network in pulpitis.
Methods
Gene expression data for m6A regulators in human pulpitis and normal pulp tissues from public GEO databases were analyzed. Bioinformatics analysis including Gene ontology (GO) functional, and Kyoto encyclopedia of genes and genomes (KEGG) pathway analyses were performed by R package, and Cytoscape software was used to study the role of m6A miRNA-mRNA regulatory network in pulpitis. Quantitative real-time PCR (qRT-PCR) was performed to validate the expression of key m6A regulators in collected human pulpitis specimens.
Results
Differential genes between pulpitis and normal groups were found from the GEO database, and further analysis found that there were significant differences in the m6A modification-related genes ALKBH5, METTL14, METTL3, METTL16, RBM15B and YTHDF1. And their interaction relationships and hub genes were determined. The hub m6A regulator targets were enriched in immune cells differentiation, glutamatergic synapse, ephrin receptor binding and osteoclast differentiation in pulpitis. Validation by qRT-PCR showed that the expression of methylases METTL14 and METTL3 was decreased, thus these two genes may play a key role in pulpitis.
Conclusion
Our study identified and validated the m6A RNA regulatory network in pulpitis. These findings will provide valuable resource to guide the mechanistic and therapeutic analysis of the role of key m6A modulators in pulpitis.
Similar content being viewed by others
Introduction
Pulpitis is a common disease in the oral cavity with a high incidence. Pulpitis is an inflammatory disease of the dental pulp which is caused by a progressive inflammatory reaction of the pulp-dentin complex caused by deep caries, trauma, etc. [1]. There is still no new breakthrough in the treatment of pulpitis at present [2]. The treatment for pulpitis is still root canal therapy to remove the pulp tissue. However, after the pulp tissue is removed from the affected tooth, there is no nutritional supply, and the brittleness increases, resulting in a decrease in the service life of the tooth. Pulpitis, as an inflammatory disease, is closely related to epigenetic modifications [3]. Research has shown that DNA methylation and RNA modification are involved in the occurrence and development of this disease [4, 5]. However, the mechanisms involved have not yet been fully elucidated, and the urgent search for the pathogenesis of pulpitis and new therapeutic targets is expected to bring good news to patients with pulpitis.
RNA in organisms can be post transcriptionally modified by more than 100 different chemical modifications, among which N6-methyladenosine (m6A) has been identified as the most abundant internal modification in eukaryotic mRNA [6]. m6 A is associated with many diseases such as cancer [7], and viral infection and inflammation are also diseases affected by m6 A [8, 9]. m6A modification is reversible and has become one of the current research hotspots. It is catalyzed by “writers” (METTL3, METTL14 and WTAP) to form a methyltransferase complex, which adds a methyl group to the N6 site of adenine in the RRACU sequence motif (R means G or A), while demethylation the enzymes ALKBH5 and FTO reverse this process, acting as an “eraser” [Full size image
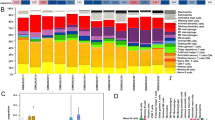
We further compared the expression of m6A-related genes in the normal group and the pulpitis group and made corresponding statistical analysis (Fig. 2). The results showed that there were significant differences in the genes ALKBH5, METTL14, METTL3, METTL16, RBM15B and YTHDF1 between normal group and the pulpitis group. It indicates that these genes may play specific roles in pulpitis.
m6A-related genes molecular interaction network and core genes interaction network
To explore the hub m6A regulators in pulpitis, we further constructed the molecular interaction network of m6A-related genes, and Cytoscape was used to extract the interaction network of core genes (Fig. 3), which further suggested that these genes RBM15B, METTL16, METTL14, YTHDF3 and ALKBH5 may be the main regulators in pulpitis m6A-modified genes.
Potential functions of genes modified by m6A regulators associated with pulpitis
To explore the role of m6A regulators in pulpitis, we further analyzed the downstream target molecules of core genes with statistical significance and intersected these molecules with differential genes in GEO (Fig. 4A), suggesting that these intersected molecules are m6A regulated molecules in pulpitis. And the number of intersected gene between GSE77459 and RBM15B targets, GSE77459 and YTHDF1 targets and GSE77459 and YTHDF3 is 110, 418, 70, respectively. We performed GO enrichment analysis and KEGG enrichment analysis at the same time (Fig. 4B-C), and the results showed that these further screened downstream genes were mainly enriched in biological processes such as cell adhesion, T cell activation, glutamatergic synapse, ephrin receptor binding, cytokine receptors, and osteoclasts differentiation. These results suggested that m6A regulators in pulpitis are mainly associated the biological processes of immune cell activation, glutamatergic synapse, cytokine activity and osteoclast differentiation, which are important for pulpitis progression.
Molecular interaction network of upstream miRNAs regulating the core m6A genes
Pulpitis is a ubiquitous chronic inflammation in the dental pulp, and miRNAs have been found to act as promoters or inhibitors of oral inflammation [17]. To explore the potential upstream regulators of hub m6A genes in pulpitis, we used the miRwalkdatabase to predict the upstream miRNAs of the core m6A genes, including has-miR-4709-3p, has-miR-5010-3p, hsa-miR-4257, hsa-miR-550b-2-5p, has-miR-7977, has-miR-29b-2-5p, has-miR-149-3p, has-miR-363-5p, has-miR-329-3p, hsa-miR-329-3p, and made a molecular interaction network (Fig. 5).
Validation of key m6A regulators in pulpitis
To further validate the key m6A regulators in pulpitis, the expression of ALKBH5, METTL3, METTL14, METTL16, RBM15B, YTHDF1, YTHDF3 and ZCCHC4 genes was detected by PCR on collected clinical tissue samples. METTL14 mRNA was confirmed to be significantly downregulated in pulpitis samples compared with controls (Fig. 6). However, YTHDF1, YTHDF3, METTL16 and METTL3 gene expression were not statistically different between the pulpitis group and the control group, and RBM15B, ZCCHC4 and ALKBH5 were up-regulated (Fig. 6). These results suggest that METTL14 may be a key regulator in pulpitis, further validating the differential expression of m6A-related genes between pulpitis and normal groups.