Abstract
Background
Chrysanthemum, one of the four major cut flowers all over the world, is very sensitive to salinity during cultivation. DNA binding with one finger (DOF) transcription factors play important roles in biological processes in plants. The response mechanism of CmDOF18 from chrysanthemum to salt stress remains unclear.
Results
In this study, CmDOF18 was cloned from Chrysanthemum morifolium, and its expression was induced by salinity stress. The gene encodes a 291-amino acid protein with a typical DOF domain. CmDOF18 was localized to the nucleus in onion epidermal cells and showed transcriptional activation in yeast. CmDOF18 transgenic plants were generated to identify the role of this gene in resistance to salinity treatment. Chrysanthemum plants overexpressing CmDOF18 were more resistant to salinity stress than wild-type plants. Under salinity stress, the malondialdehyde content and leaf electrolyte conductivity in CmDOF18-overexpressing transgenic plants were lower than those in wild-type plants, while the proline content, chlorophyll content, superoxide dismutase activity and peroxidase activity were higher than those in wild-type plants. The opposite findings were observed in gene-silenced plants compared with wild-type plants. The gene expression levels of oxidoreductase increased in CmDOF18-overexpressing transgenic plants but decreased in CmDOF18-SRDX gene-silenced transgenic plants.
Conclusion
In summary, we analyzed the function of CmDOF18 from chrysanthemum, which may regulate salinity stress in plants, possibly due to its role in the regulation of oxidoreductase.
Similar content being viewed by others
Background
Transcription factors (TFs) activate or inhibit target gene transcription by directly binding to cis-regulatory elements of promoters acting as gene expression regulators [1, 2]. The DNA binding with one finger (DOF) family is a classic protein in the Cys2-His2 zinc finger superfamily of TFs [3]. The DOF TFs contain a single conserved zinc finger motif named the DOF domain with a Cys2-His2 zinc finger containing 50–52 amino acid residues that binds to a specific element with 5’-AAAG-3’ sequences [4, 5]. In recent years, numerous members of the DOF TF family have been reported in a diverse variety of plants [S6), suggesting that CmDOF18 might play a role in the plant salinity stress response by modulating the expression of oxidoreductase. The expression levels of the abovementioned DEGs were verified by qRT-PCR (Fig. 6).
Discussion
Structural characteristics and transcriptional activation activity of CmDOF18
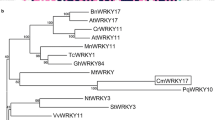
DOF TFs are characterized mainly by the presence of the conserved DOF DNA-binding domain in the N-terminus and a C-terminal transcriptional activation domain [3, 43, 44]. In this study, CmDOF18, a group VI DOF gene, was obtained from chrysanthemum. Sequence analysis showed that it contains a highly conserved DOF domain (Fig. 1A), suggesting that CmDOF18 might be able to bind to the element with the sequence 5’-AAAG-3’. DOF TFs contain a bipartite nuclear localization signal (NLS) that partly overlaps with the conserved DOF DNA-binding domain [45, 46], and the subcellular localization of CmDOF18 showed that it localized to the nucleus. Transactivation assays showed that CmDOF18 is transcriptionally active, suggesting that it might activate the expression of downstream genes to exert its effects.
CmDOF18 positively regulates the resistance of chrysanthemum to salinity treatment
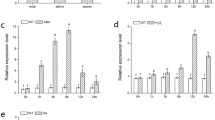
Several results have previously shown that DOF family members play important roles in resistance to various abiotic stresses in plants [6, 13, 47,48,49,50,51]. VyDOF8 expression is significantly induced by cold treatment, drought treatment, and salt treatment, and VyDOF8-overexpressing tobacco shows enhanced drought tolerance due to increases in abscisic acid and promotion of stress-responsive gene expression [22]. OsDOF15 can bind to the DOF motif in the downstream OsACS1 promoter and may participate in primary root elongation under salt stress by regulating cell proliferation in the root meristem, via restriction of ethylene biosynthesis [24]. RNA interference (RNAi) of SlDof22 in transgenic lines increases ascorbic acid (AsA) levels and affects the expression of genes in the D-mannose/L-galactose pathway and AsA recycling, resulting in reduced tolerance to salt stress by significantly downregulating the SlSOS1 gene [52]. In the present study, CmDOF18 expression was significantly induced by salinity stress (Fig. 2A), and plants overexpressing CmDOF18 exhibited improved resistance to salinity stress, while gene-silenced plants showed reduced resistance to salt stress.
Various physiological indices will change under salt stress, such as malondialdehyde, leaf relative conductivity, proline, chlorophyll, SOD, POD, and the degree of these indices can indicate the strength of plant resistance to salt stress [53,54,55]. MDA, electrolyte leakage, proline, and chlorophyll concentrations are generally used as indicators of plant membrane damage levels under salt stress [31, 56]. As MDA is a product of membrane lipid peroxidation, MDA content can serve as an indicator of the degree of cellular membrane lipid peroxidation occurring as a response to stress [57]. In addition, electrolyte leakage reflects membrane injury caused by stresses [58]. The MDA content and leaf relative electrolyte conductivity in CmDOF18-overexpressing lines were lower than those in WT and gene-silenced lines under salt stress, suggesting that CmDOF18 improved plant salinity tolerance by maintaining the membrane integrity of plants. Proline functions as an osmotic protectant for various cellular structures during episodes of abiotic stress, and its content strongly increases in response to a variety of stresses in plants, such as drought, salt stress, cold injury, etc. [59]. In present study, when the plants suffers from salt stress, the contents of proline showed an increased trend compared with that under non-stress, not only in wild-type plants and CmDOF18-overexpressing transgenic plants, but also in CmDOF18-SRDX gene-silenced transgenic plants, suggesting that plants could accumulate proline, acting as an osmotic regulator, to regulate the response to salt stress, which is entirely consistent with previous reports [60]. Further analysis showed that more proline accumulated in the leaves of CmDOF18-overexpressing lines than that in WT, lower proline accumulated in gene-silenced lines compared with that in WT under salt stress, suggesting that CmDOF18 might improve plant salinity tolerance potentially by accumulation of proline. Chlorophyllase activity increases under salt stress, leading to decreased chlorophyll content [61]. In our study, we found that the content of chlorophyll decrease in plants under salt stress, and higher content in CmDOF18-overexpressing lines, lower content in gene-silenced lines than that in wild-type plants, meaning that CmDOF18 might response to salt stress by regulating synthesis or degradation of chlorophyll. Oxidative damage is caused by the accumulation of reactive oxygen species (ROS), which occurs under various stresses in plants [30]. The two enzymes SOD and POD are involved in oxidative protection [62]. SOD catalyzes O2− to produce oxygen and H2O2 by catalyzing the dismutation reaction, and POD metabolizes H2O2 to H2O through synergistic action. Consistent with this result, the SOD and POD activity under salinity stress was higher in CmDOF18-overexpressing lines than in WT and gene-silenced lines, suggesting that these enzymes contribute to improving the resistance to salinity stress in CmDOF18-overexpressing lines. These results suggested that CmDOF18 genes might resist salinity stress by regulating lipid peroxidation, osmoregulatory substance, and ROS accumulation in plants.
CmDOF18-altered salt resistance is potentially related to oxidoreductase
DOF proteins can respond to salt stress by regulating a variety of pathways, including osmotic substance synthesis, protective enzyme synthesis, Na + excretion, and so on [63, 64]. ThDof1.4 could increase the proline level and enhance ROS scavenging capability to improve salt and osmotic stress tolerance in Tamarix hispida [48]. TaZNF, a wheat DOF protein, significantly improved salt tolerance by controlling the expression of many downstream genes to increase Na + excretion in Arabidopsis [49]. Here, transcriptome analysis showed that the identified differently expressed genes between wild-type and CmDOF18 transgenic plants are mainly involved in oxidoreductase activity (Table S6). Oxidoreductase is an enzyme that catalyzes oxidation-redution reactions, which exist widely in organisms. The main function of oxidoreductase is to produce energy and synthesize various substances needed for plant growth and the interaction between plants and the environment. Cytochromes P450s (P450s) are a large superfamily of heme-containing monooxygenases, that function in metabolic detoxification and participate primarily in the synthesis of plant secondary metabolites and in plant defense [65]. The expression of PtCYP714A3, a cytochrome P450 monooxygenase gene, is greatly induced by salt and osmotic stress in plants, and transgenic rice plants exhibit enhanced tolerance to salt and maintained more Na+ in both shoot and root tissues under salinity stress than WT plants, suggesting that PtCYP714A3 plays a crucial role in shoot responses to salt toxicity in rice by regulating gibberellin synthesis [66]. Alcohol dehydrogenases (ADHs) in plants are encoded by a multigene family, which participates in growth, development, and adaptation in many plant species. ScADH3, which maintains the steady state of ROS by regulating ROS-related genes, is also related to cold tolerance in transgenic tobacco, as indicated by functional analysis [67]. In plants, lipoxygenases (LOXs) are involved in various physiological processes, including defense responses to biotic and abiotic stresses. CaLOX1 plays a crucial role in plant stress responses by modulating ABA- and stress-responsive marker gene expressions, lipid peroxidation and H2O2 production [68]. CmLOX10 positively regulates drought tolerance through jasmonic acid -mediated stomatal closure in oriental melon [69]. In the present study, we found that the expression of six oxidoreductase genes, including CmCYP71A1, CmCYP1, CmCYP2 and CmCYP3 encoding cytochrome P540 monooxygenase; CmADH1 encoding an alcohol dehydrogenase; and CmLOX1 encoding a lipoxygenase, was increased in CmDOF18-overexpressing plants but decreased in SRDX lines. Thus, we propose that CmDOF18 mediates resistance to salinity stress and that the mechanism could be related to oxidoreductases such as cytochrome P450 monooxygenases, alcohol dehydrogenases, and lipoxygenases. However, the specific mechanism remains to be clarified, and more data are needed before a definitive conclusion can be made.
Hypothetical model for CmDOF18 function during salt stress. Accumulation of CmDOF18 during salinity stress results in an increase in oxidoreductase gene expression. Genes shown dotted box are those oxidoreductase genes (CmCYP71A1, CmCYP1, CmCYP2, CmCYP3, CmADH1, and CmLOX1) that responded to salt stress
Conclusion
In summary, CmDOF18 was cloned from chrysanthemum, and its expression was induced by salinity stress, indicating that CmDOF18 mediates resistance to salinity stress in chrysanthemum. The expression levels of oxidoreductase genes (CmCYP71A1, CmCYP1, CmCYP2, CmCYP3, CmADH1, and CmLOX1) increased in CmDOF18-overexpressing plants but decreased in CmDOF18-SRDX gene-silenced plants. It appeared that CmDOF18 activates the above genes in CmDOF18-overexpressing lines during salt stress, which therefore results in tolerance to salinity (Fig. 7).
Data availability
The raw RNAseq data has been successfully uploaded to NCBI and the accession number for our submission is: PRJNA1037609. The materials used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DOF:
-
DNA binding with one finger
- TFs:
-
Transcription factors
- ORF:
-
Open reading frame
- MDA:
-
Malondialdehyde
- SOD:
-
Superoxide dismutase
- POD:
-
Peroxidase
- FPKM:
-
Fragments per kilo base per million
- WT:
-
Wild-type
- EAR:
-
ERF-associated amphiphilic repression
- DEGs:
-
Differentially expressed genes
- RNAi:
-
RNA interference
- ROS:
-
Reactive oxygen species
- P450s:
-
Cytochromes P450s
- ADHs:
-
Alcohol dehydrogenases
- LOXs:
-
Lipoxygenases
References
de Pater S, Greco V, Pham K, Memelink J, Kijne J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996;24(23):4624–31.
Kreps J, Wu Y, Chang H, Zhu T, Wang X, Harper J. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130(4):2129–41.
Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7(12):555–60.
Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T. Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA. 2004;101(20):7833–8.
Gupta S, Malviya N, Kushwaha H, Nasim J, Bisht N, Singh V, et al. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta. 2015;241(3):549–62.
Dong C, Hu H, **e J. Genome-wide analysis of the DNA-binding with one zinc finger (dof) transcription factor family in bananas. Genome. 2016;59(12):1085–100.
Li H, Huang W, Liu Z, Wang Y, Zhuang J. Transcriptome-based analysis of Dof family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). International Journal of Genomics. 2016; 2016: 5614142.
Song A, Gao T, Li P, Chen S, Guan Z, Wu D, et al. Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front Plant Sci. 2016;7:199.
Chen M, Liu X, Huan L, Sun M, Liu L. Genome-wide analysis of Dof family genes and their expression during bud dormancy in peach (Prunus persica). Sci Hort. 2017;214:18–26.
Yang Q, Chen Q, Zhu Y, Li T. Identification of MdDof genes in apple and analysis of their response to biotic or abiotic stress. Funct Plant Biol. 2017;45(5):528–41.
Fang Z, Jiang W, He Y, Ma D, Liu Y, Wang S, et al. Genome-wide identification, structure characterization, and expression profiling of Dof transcription factor gene family in wheat (Triticum aestivum L). Agronomy. 2020;10(2):294.
Khan I, Khan S, Zhang Y, Zhou J. Genome-wide analysis and functional characterization of the Dof transcription factor family in rice (Oryza sativa L). Planta. 2021;253:1–14.
Liu W, Ren W, Liu X, He L, Qin C, Wang P, et al. Identification and characterization of dof genes in Cerasus Humilis. Front Plant Sci. 2023;14:1152685.
Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol. 2003;3(1):17.
Guo Y, Qiu L. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE. 2013;8(9):e76809.
Park D, Lim P, Kim J, Cho D, Hong S, Nam H. The Arabidopsis COG1 gene encodes a dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 2003;34(2):161–71.
Isabel-LaMoneda I, Diaz I, Martinez M, Mena M, Carbonero P. Sad: a new dof protein from barley that activates transcription of a cathepsin b-like thiol protease gene in the aleurone of germinating seeds. Plant J. 2003;33(2):329–40.
Xu J, Dai H. Brassica napus Cycling Dof factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul. 2016;80(3):315–22.
Fornara F, Panigrahi K, Gissot L, Sauerbrunn N, Rühl M, Jarillo J, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17(1):75–86.
Negi J, Moriwaki K, Konishi M, Yokoyama R, Nakano T, Kusumi K, et al. A dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr Biol. 2013;23(6):479–84.
Ruta V, Longo C, Lepri A, De Angelis V, Occhigrossi S, Costantino P, et al. The DOF transcription factors in seed and seedling development. Plants (Basel). 2020;9(2):218.
Li G, Xu W, **g P, Hou X, Fan X. Overexpression of VyDOF8, a Chinese wild grapevine transcription factor gene, enhances drought tolerance in transgenic tobacco. Environ Exp Bot. 2021;190(1):104592.
Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. The dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2010;61(2):312–23.
Qin H, Wang J, Chen X, Wang F, Peng P, Zhou Y, et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019;223(2):798–813.
Ravindran P, Verma V, Stamm P, Kumar P. A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol Plant. 2017;10(10):1307–20.
Yang Y, He Z, Bing Q, Duan X, Chen S, Zeng M, et al. Two Dof transcription factors promote flavonoid synthesis in kumquat fruit by activating C-glucosyltransferase. Plant Science: Int J Experimental Plant Biology. 2022;318:111234.
Venkatesh J, Park S. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (dof) transcription factor family in potato. Plant Physiol Biochem. 2015;94:73–85.
Wu Z, Cheng J, Cui J, Xu X, Liang G, Luo X, et al. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L). Front Plant Sci. 2016;7:574.
Zhang L, Liu B, Zheng G, Zhang A, Li R. Genome-wide characterization of the SiDof gene family in foxtail millet (Setaria italica). BioSystems. 2017;151:27–33.
Zhu W, Jiang J, Chen S, Wang L, Xu L, Wang H, et al. Intergeneric hybrid between Chrysanthemum × morifolium and Artemisia Japonica achieved via embryo rescue shows salt tolerance. Euphytica. 2013;191(1):109–19.
Deinlein U, Stephan A, Horie T, Luo W, Xu G, Schroeder J. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19(6):371–9.
Subramanian L. Primer premier 5. Biotechnol Softw Internet Journal: Comput Softw J Scient. 2000;1:270–2.
Tamura K, Stecher G, Kumar S. Mega11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7.
Li P, Song A, Gao C, Wang L, Wang Y, Sun J, et al. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015;34(8):1365–78.
Li P, Song A, Gao C, Jiang J, Chen S, Fang W, et al. The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol Biochem. 2015;95:26–34.
An J, Song A, Guan Z, Jiang J, Chen F, Lou W, et al. The over-expression of Chrysanthemum Crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol Biol Rep. 2014;41(6):4155–62.
Kim D, Langmead B, Salzberg S. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60.
Song A, Su J, Wang H, Zhang Z, Zhang X, Van de Peer Y, et al. Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated chrysanthemum. Nat Commun. 2023;14:2021.
Langmead B, Salzberg S. Fast gapped–read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9.
Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13(8):1959–68.
Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the ERF motif, a repression domain in Arabidopsis. Plant J Cell Mol Biology. 2003;34(5):733–9.
Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. Identification of the minimal repression domain of superman shows that the dlelrl hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun. 2004;321(1):172–8.
Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem. 1993;268(21):16028–36.
Yanagisawa S, Schmidt R. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999;17(2):209–14.
Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004;45(4):386–91.
Ravel C, Nagy I, Martre P, Sourdille P, Dardevet M, Balfourier F, et al. Single nucleotide polymorphism, genetic map**, and expression of genes coding for the DOF wheat prolamin-box binding factor. Funct Integr Genom. 2006;6(4):310–21.
He L, Su C, Wang Y, Wei Z. ATDOF5.8 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie. 2015;110:17–24.
Zang D, Wang L, Zhang Y, Zhao H, Wang Y. ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix Hispida. Plant Mol Biol. 2017;94(4–5):495–507.
Ma X, Liang W, Gu P, Huang Z. Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiol Biochem. 2016;106:129–40.
Zhou Y, Cheng Y, Wan C, Li J, Chen J. Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ. 2020;8(1–2):e8358.
Zou X, Sun H. Dof transcription factors: specific regulators of plant biological processes. Front Plant Sci. 2023;14:1044918.
Cai X, Zhang C, Shu W, Ye Z, Li H, Zhang Y. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem Biophys Res Commun. 2016;474(4):736–41.
EI-Bassiouny H, Bekheta M. Effect of salt stress on relative water content, lipid peroxidation, polyamines, amino acids and ethylene of two wheat cultivars. Int J Agric Biology. 2005;7(3):363–8.
Wang X, Wang H, Wu F, Liu B. Effects of cinnamic acid on the physiological characteristics of cucumber seedlings under salt stress. Front Agric China. 2007;1:58–61.
Yu J, Chen S, Zhao Q, Wang T, Yang C, Diaz C, et al. Physiological and proteomic analysis of salinity tolerance in Puccinellia tenuiflora. J Proteome Res. 2011;10(9):3852–70.
Zhu J. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53(1):247.
Zhu J. Plant salt tolerance. Trends Plant Sci. 2001;6(2):66–71.
Sun X, Li Y, Cai H, Bai X, Ji W, Ding X, et al. The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses. J Plant Res. 2012;125(3):429–38.
Parida A, Das A. Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf. 2005;60(3):324–49.
Misra N, Gupta A. Effect of salt stress on proline metabolism in two high yielding genotypes of green gram. Plant Sci. 2005;169(2):331–9.
Santos C. Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Sci Hort. 2004;103(1):93–9.
Jaiti F, Verdeil J, Hadrami I. Effect of jasmonic acid on the induction of polyphenoloxidase and peroxidase activities in relation to date palm resistance against Fusarium oxysporum f. sp. albedinis. Physiological Mol Plant Pathol. 2010;74(1):84–90.
Noguero M, Atif R, Ochatt S, Thompson R. The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Sci. 2013;209:32–45.
Li Y, Tian M, Feng Z, Zhang J, Lu J, Fu X, et al. GhDof1.7, a Dof transcription factor, plays positive regulatory role under salinity stress in upland cotton. Plants (Basel). 2023;12(21):3740.
Finnigan J, Young C, Cook D, Charnock S, Black G. Cytochromes p450 (p450s): a review of the class system with a focus on prokaryotic P450s. Adv Protein Chem Struct Biology. 2020;122:289–320.
Wang C, Yang Y, Wang H, Ran X, Li B, Zhang J, et al. Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol J. 2016;14(9):1838–51.
Su W, Ren Y, Wang D, Su Y, Que Y. The alcohol dehydrogenase gene family in sugarcane and its involvement in cold stress regulation. BMC Genomics. 2020;21(1):521.
Lim C, Han S, Hwang I, Kim D, Hwang B, Lee S. The pepperl lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 2015;56(5):13.
**ng Q, Liao J, Cao S, Li M, Qi H. CmLOX10 positively regulates drought tolerance through jasmonic acid -mediated stomatal closure in oriental melon (Cucumis melo var. Makuwa Makino). Sci Rep. 2020;10(1):17452.
Acknowledgements
We are grateful to Professor **g Sun (Yangzhou University) for helpful suggestion. We also thank Professor Ai** Song (Nan**g Agricultural University) for providing the pMDC43 vector and p35S-SRDX vector.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 32002083), the Natural Science Foundation of Jiangsu Province (BK20210163), the Foundation of Central Laboratory of **nyang Agriculture and Forestry University (Grant No. FCL202002) and the Program for Innovative Research Team of Horticultural Plant Resources and Utilization in **nyang Agriculture and Forestry University (XNKJTD-012).
Author information
Authors and Affiliations
Contributions
All authors contributed largely to the work presented in this article. Conceived and designed the experiment: L. P. L. and W. Z. Y. Performed the experiments: C. X. R., F. T. T., C. J. J. Language modification: Y. J. H. Wrote the paper: L. P. L. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
In this study, the chrysanthemum plants used were from College of Horticulture, **nyang Agriculture and Forestry University. These plant materials do not include any wild species at risk of extinction. No specific permits were required for sample collection in this study. We complied with relevant institutional, national, and international guidelines and legislation for plant study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, P., Fang, T., Chong, X. et al. CmDOF18 positively regulates salinity tolerance in Chrysanthemum morifolium by activating the oxidoreductase system. BMC Plant Biol 24, 232 (2024). https://doi.org/10.1186/s12870-024-04914-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-024-04914-y