Abstract
Phyllodistomum is the large digenean group of fish parasites, with 25 species described so far in the Indian subcontinent. Here, we redescribed two adult species of Phyllodistomum (P. srivastava Rai 1964 and P. parorchium Jaiswal 1957) collected from freshwater fish Heteropneustes fossilis Bloch, 1974 and Glossogobius giuris Ham, 1822, respectively, and an unknown Phyllodistomum metacercaria from shrimp (Macrobrachium dayanum Henderson, 1893). These parasites were genetically characterized using 28S and first and second internal transcribed spacers (ITS1 and ITS2) regions of the nuclear ribosomal DNA and CoxI region of the mitochondrial (mt) DNA to establish the link between metacercaria and adult. Morphologically, both the unknown metacercaria in shrimp and adult Phyllodistomum srivastava in fish, resembled in terms of crenulated margin of hind body, arrangement of diagonal testes, bipartite seminal vesicle, and compact paired vitelline masses. The two adult parasite species, P. srivastava from P. parorchium, were different in terms of shape and size of the body, ratio of suckers, the absence of crenulated margin of hind body, a single chambered seminal vesicle, and deeply lobed paired vitelline masses, in the former species. Comparison of the 28S, ITS, and mtCoxI sequence data suggested P. srivastava and Phyllodistomum metacercaria belong to the same species, and supported the distinction between P. srivastava and P. parorchium. Exploring the potential impact of Phyllodistomum infection on host behaviour and health would be prospective areas for future research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Phyllodistomum Braun 1899 is a large group of digenean fish parasites, comprising 120 species, under the family Gorgoderidae, Looss 1901. Phyllodistomum spp. are known to parasitize both marine and freshwater fishes, with occasional reports of infestations in amphibians (Campbell 2008). They have global distribution. The genus Phyllodistomum is characterized by broad, tapering fore body and foliate hind body with more or less crenulated body margin (Campbell 2008). Of the 120 species belonging to the genus Phyllodistomum, 25 nominal species, distinguished by conventional morphological characters, are so far reported from freshwater fish in the Indian subcontinent (Table 1) (Jaiswal 1957; Rai 1964; Sarwat 2011; Naz and Siddique 2012; Sen 2014). In previous studies, Pandey (1970) and Rai (1964) collected phyllodistome metacercariae encysted in the hepatopancreas of crustaceans, Macrobrachium dayanum Henderson (1893) and obtained their adults, by performing feeding experiments in fish, Heteropneustes fossilis. They identified these phyllodistomes as P. srivastava and P. lucknowensis.
Digeneans commonly undergo a series of developmental stages involving different hosts, making their life cycles intricate and challenging to study solely through traditional methods (Aghlmandi et al. 2018). Sequence data have proven to be invaluable in elucidating the life cycles of digeneans, which typically exhibit an indirect life cycle. The use of genetic information, such as DNA sequencing of specific regions like 28S rRNA, ITS1, and CoxI, has provided crucial insights into the complex life cycles of these parasitic flatworms (Huston et al. 2018; Rochat et al. 2020; Shamsi et al. 2021a; Shamsi et al. 2021b; Shamsi et al. 2023, Barton et al. 2022; Choudhary et al. 2022).
Cribb (1987) highlighted that the proper identification of Phyllodistomum spp. is challenging due to significant intra-specific morphological variations and inadequate descriptions of several species. However, recent advances in molecular biology, particularly PCR and sequencing-based molecular techniques, have proven effective in identifying and distinguishing digenean parasites (Blair et al. 1996; Hust et al. 2004; Goswami et al. 2009). These molecular techniques have been particularly useful in elucidating the taxonomic status of Phyllodistomum species from various parts of the world. For instance, molecular studies have successfully identified and distinguished species such as P. centropomi, P. lacustri, P. inecoli, P. cribbi, P. wallacei, and P. spinopapillatum from Mexico; P. kanae, P. parasiluri, and P. elongatum from Japan; P. folium from Spain; P. pseudofolium and P. angulatum from Lithuania and Russia; P. magnificum and P. symmetrochis from Australia; and P. staffordi and P. brevicaceum from Canada (Mendoza-Garfias and de León 2005; Peribáñez et al. 2011; Cutmore et al. 2013; Razo-Mendivil et al. 2013; de León et al. 2015a; de León et al. 2015b; Nakao 2015; Urabe et al. 2015; Stunžėnas et al. 2017). However, the molecular study of Phyllodistomum species from India remains entirely unexplored. The ribosomal DNA (rDNA) is helpful in resolving the phylogeny because it is universal and composed of highly conserved as well as variable domains (Tkach et al. 2000; Mwita and Nkwengulila 2010). The internal transcribed spacer regions (ITS1 and ITS2) also include a high degree of inter- and intra-specific genetic variations (Nolan and Cribb 2005; Littlewood 2008; Choudhary et al. 2015). Additionally, the mitochondrial cytochrome oxidase 1 (Cox1) has also been used for distinguishing digenean species and inferring phylogenies (Georgieva et al. 2013). Therefore, by coupling molecular and morphological approaches, uncertainties of existing species could be resolved. Also, the preliminary identification and molecular data allow establishing a link between metacercaria found in shrimp and adult in freshwater fish, not yet elucidated in India.
We hereby present first molecular data to (1) distinguish and identify two Phyllodistomum species and (2) correlate one with its metacercaria, in combination with sequences, and (3) to determine the systematic position.
Materials and methods
Parasite collection and identification
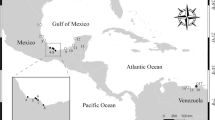
During winter months (November to January 2016), 65 live specimens of freshwater fish and shrimp were captured from the Gomti River, Lucknow, India (Fig. 1). To detect adult Phyllodistomum infection, the fish were euthanized through spinal severance and then examined under a stereomicroscope. In addition, numerous specimens of naturally infected Phyllodistome metacercariae from the hepatopancreatic tissue of the freshwater shrimp were collected. This work was conducted under animal ethic approval 20/I/2023/IAEC/LU.
Morphological study
The recovered worms were examined under a light microscope and identified using references from the literature (Yamaguti 1934; Pandey and Agrawal 2013; Pandey and Agrawal 2018). Once identified, they were fixed in 70% ethanol for whole mount preparations, and a small piece was preserved in absolute ethanol for DNA extraction. Permanent mounts were prepared by staining the fixed whole mounts in aceto-alum carmine. Subsequently, the specimens were dehydrated through a graded ethanol series (50%, 70%, 90%, and absolute ethanol), cleared in clove oil, and then mounted on glass slides using DPX mounting medium. Figures were illustrated by drawing tube, attached to a phase-contrast light microscope (Olympus BX-51). All morphometric measurements (in millimeters) were taken with the aid of ocular micrometer. Voucher specimens were deposited at Helminthological Collection of the Zoological Survey of India, Kolkata.
Molecular study
DNA was extracted using Qiagen’s DNeasy Blood and Tissue Kit according to the manufacturer protocols. Three nuclear ribosomal loci (28S, ITS1, ITS2) and one mitochondrial cytochrome oxidase subunit I (CoxI) were amplified using the following primer sets: 28S (Forward): 5′-ACCCGCTGAATTTAAGCAT-3′ and (Reverse): 5′-CTCTTCAGAG TACTTTTCAA-3′; ITS1 (Forward): 5′-GTCGTAACAAGGTTTCCGTA-3′ and 5′-TCTAGATGCGTTCGA(G/A)TGTCGATG-3′; ITS2 3S (Forward): 5′-GGTACCGTGGATCACTCGGCTCGTG-3′ and A28 (Reverse): 5′-GGGATCCTGGTTAGTTTCTTTTCCTCCGC-3′; and COI JB3 (Forward): 5′-TTTTTTGGGCATCCTGAGGTTTAT-3′ and JB4.5 (Reverse): 5′-TAAAGAACATAATGAAAATG-3′ primer (Bowles et al. 1993; Mollaret et al. 2000; Prasad et al. 2008). Each PCR amplification reaction is performed in a final volume of 12.5 μl, containing 10X buffer (100 mM Tris, pH 9.0), 50 mM KCl and 15mM MgCl2, 2.5 U Taq polymerase enzyme, 10 mM of each deoxynucleotide triphosphates (dNTPs), and 3μl DNA. The PCR conditions are as follows: initial denaturation at 94 °C for 5 min, annealing for 28S at 54 °C (1 min), ITS-1 at 54 °C (1.10 min), ITS-2 at 57 °C (1.10 min), mt CoxI at 56 °C (1.10 min), and final extension at 72 °C for 10 min. PCR products are checked on 2% agarose gel in TAE buffer, stained with ethidium bromide (EtBr) and visualized under UV light. The acquired PCR products were purified and subjected to Sanger sequencing in the forward direction, using an ABI3730xl DNA Analyzer (Applied Biosystems, Foster City, CA).
Similarity search analysis of nucleotide sequence was performed by Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast) and Clustal W (http://www.ebi.ac.uk/clustalw/) used for multiple sequence alignment. BioEdit software version 7.0.9.0 (Hall 1999) was used for sequence identities.
Phylogenetic trees were constructed (Table 2) using MEGA 11 (Tamura et al. 2013). Neighbor-joining (NJ) and maximum likelihood (ML) analysis were performed for each data set (28S, ITS1, ITS2, and mtCoxI). The best nucleotide substitution modal was estimated “Kimura 2-parameter model” for NJ tree and “General time reversible model” for ML trees with a gamma distribution of rates and proportion of invariant sites (GTR+G+I). The reliability of internal branches in all trees was evaluated by using the bootstrap method, 1000 replicates.
Results
Based on their morphological features, the specimens of Phyllodistomum were identified as P. srivastava, P. parorchium, and one unknown Phyllodistomum metacercaria (Fig. 2). Details of the parasites found in the present study are provided in Table 3.
Phyllodistomum spp. found in the present study: A Adult P. srivastava, B detail of male and female reproductive complex of adult P. srivastava, C adult P. parorchium, D detail of male and female reproductive complex of adult P. parorchium, E metacercaria of Phyllodistomum sp. Note hind body with more or less crenated margin, elongated narrow esophagus, lobed testes positioned asymmetrically, and ovary with smooth margin in P. srivastava versus hind body with broad less crenulated margin, tubular esophagus, deeply lobed testes positioned almost symmetrically and lobulated ovary in P. parorchium
Morphological redescriptions
Measurements of the specimens found in the present study (n = 10) are given in millimeters in Table 4.
P. srivastava Rai, 1964 (Fig. 1a)
Description: Body aspinose, elongated, pyriform, hind body with more or less crenated margin. Oral sucker globular, subterminal or terminal, pharynx absent, esophagus straight, thin, long, bifurcates into two intestinal ceca, extending up to hind end of the body. Ventral sucker larger then oral sucker situated in the middle of the body. Testes deeply lobed, oblique, intercecal, post-equatorial, tandem, vas deferens leading to bipartite vesicula seminalis, ejaculatory duct short, inconspicuous, opening into the genital atrium, cirrus lacking. Ovary subglobular or lobed, intercecal, anterior to left testis. Vitellaria paired compact masses, slightly lobed, round or oval, between ventral sucker and ovary. Laurer’s canal not observed. Mehlis’ gland small, between vitelline masses. Uterus extensively coiled, occupying entire hind body, intercecal or extracecal, occupying posterior half of hind body, uterine coils extending posteriorly beyond ceca, filled with eggs, genital pore sub-median. Excretory bladder tubular, opening by a median excretory aperture in the caudal notch. Eggs small, operculated.
P. parorchium Jaiswal, 1957 (Fig. 1b)
Description: Body aspinose, less crenulated margin, broad, oral sucker globular, subterminal or terminal, ventral sucker rounded, equatorial, larger than an oral sucker, pharynx absent, esophagus tubular, leading into two broad intestinal ceca, extending up to the hind region of the body. Testes two, deeply lobed, oblique, intercecal, anterior testis situated close to vitellarium, posterior testis close to ovary, vesicula seminalis free in the parenchyma, opening by a short duct at genital pore. Cirrus sac absent, seminal vesicle single chambered, just posterior to intestinal bifurcation, before opening into genital atrium, anterior end surrounded by prostatic cells. Ejaculatory duct short. Genital pore median at the level of intestinal bifurcation, sperms visible in the uterus. Ovary trilobed, anterior and oblique to posterior testis. Vitelline massed two, deeply lobed with irregular margins, behind ventral sucker. Mehlis’ gland small, between vitelline masses. Laurer’s canal well developed. Uterus highly coiled, intercecal or extracecal, descending limb of uterus occupying posterior half of hind body, ascending limb of uterus running forward dorsal to ventral sucker and narrows to form metraterm which opens into genital pore. Excretory bladder tubular, run in a zig-zag manner, up to ventral sucker, expanding into a sac-like structure and excretory pore situated at the posterior end of the body. Eggs oval, thin, light brown and operculated.
Metacercaria of P. srivastava Rai, 1964 (Fig. 1c)
Description: Body elongated, aspinose, pyriform, oral sucker globular, subterminal or terminal, ventral sucker equatorial, larger than an oral sucker, pharynx absent, esophagus straight, long, bifurcates into two broad intestinal caeca, extending up to the hind end of the body. Testes two, lobed, intercecal, oblique, almost similar size, in posterior third of hind body, vas deferens leading to bipartite seminal vesicle, ejaculatory duct short, inconspicuous, opening into the genital atrium, cirrus lacking. Genital pore situated slightly away from intestinal bifurcation. Ovary subglobular or lobed, behind left or right vitellarium. Vitelline masses paired, compact, slightly lobed, posterior to ventral sucker, Mehlis’ gland dorsal, posterior to ventral sucker, between vitelline masses. Laurer’s canal not observed. Excretory vesicle long, tubular, lies between testes, opening by the median excretory aperture.
Molecular results
Sequences of the 28S (374-384 bp long), ITS1 (973-982 bp long), ITS2 (433-454 bp long), and CoxI (388-433 bp long) were obtained successfully for all taxa in the present study. The nucleotide sequences obtained in this study were submitted to NCBI/GenBank database (Table 2).
Alignment and comparison of the sequences between the metacercaria in the present study and adult P. srivastava revealed 0.6%, 0.0%, 0.0%, and 0.2% bp difference in 28S, ITS1, ITS2, and Cox1 regions, respectively. The bp difference for these regions, between metacercaria in the present study and adult P. parorchium, were 2.7%, 3.8%, 3.7%, and 3.7%, respectively, which was within the range observed for the bp difference between distinct species, P. srivastava and P. parorchium, 2.9%, 4.0%, 3.8%, and 6.3, respectively.
In the phylogenetic trees built based on these regions (Fig. 3), metacercaria found in the present study consistently grouped with P. srivastava, with P. parorchium grou** closely but distinctly from them.
Neighbor joining and maximum likelihood phylogenetic trees built based on the sequences of A 28S, B ITS-1, C ITS2, and D Mt CoxI gene of Phyllodistomum spp. (P. srivastava, P. parorchium) and one Phyllodistomum metacercaria using the member of the genus Phyllodistomum. The number preceding the GenBank accession numbers for their 28S r RNA gene sequences. The numbers of the internodes are NJ bootstrap values (above) and ML bootstrap values (below)
Discussion
The taxonomic status of two species of Phyllodistomum in India was investigated through a comprehensive analysis combining morphology and molecular techniques. The morphological characteristics of the specimens closely resembled the original description of P. srivastava, in terms of the ratio of suckers, diagonal arrangement of testes, bipartite vesicula seminalis, and paired vitellaria (Table 4). However, some minor differences in body shape were observed, which could potentially be attributed to variations in fixation and mounting techniques used during the study. A consistent finding in our measurements was that the specimens appeared smaller compared to those documented by Rai (1964). Additionally, Rai (1964) reported presence of papillae in live specimen which gradually disappeared after fixation. This observation underscores the importance of considering the impact of fixation on morphological features, as it could lead to changes or loss of certain characteristics, potentially affecting the accuracy of taxonomic comparisons.
Cribb (1987) described Pseudophyllodistomum johnstoni which included five previously described species of Phyllodistomum. These species were reclassified under the new genus as follows: P. macrobrachicola (Yamaguti 1934) comb. nov., P. lesteri (Wu, 1938) comb. nov., P. srivastava (Rai 1964) comb. nov., P. lucknowense (Pandey 1970) comb. nov., and P. mingense (Tang, 1985) comb. nov. However, there are clear distinguishing features between P. johnstoni and P. srivastava. P. johnstoni which can be readily distinguished from P. srivastava by shape and size of body, its nearly equal oral and ventral sucker, single chambered seminal vesicle, and saccular excretory bladder. Additionally, P. johnstoni occur as a parasite of the urinary bladder of the Australian freshwater fish Leiopotherapon unicolor, whereas the P. srivastava recovered from the Indian cat fish Heteropneustes fossilis (Bloch 1974).
In case of P. parorchium, the present specimens closely resembled the description given by Jaiswal (1957) in terms of the ratio of suckers, position of genital pore, shape of testes, ovary, and vitellaria (Table 4). However, there are significant distinguishing traits between P. srivastava and P. parorchium. These characteristics include shape and size of body, absence of a crenulated margin on the hind body, presence of a large ventral sucker, a single chambered seminal vesicle, and deeply lobed vitellarium. Pandey (1970) described metacercaria od P. lucknowense, which was found in the same host, Macrobrachium dayanum (Henderson). The metacercaria P. lucknowense differs from P. srivastava in several aspects, such as the presence of a crenulated body margin, a spinose body, a saccular vesicula seminalis, and shape of the testes. Additionally, the present specimens of P. srivastava also differ from P. lucknowense Pandey 1970 by spinose body, saccular vesicula seminalis, and size of eggs. So far, only two metacercariae of Phyllodistomum, namely, P. srivastava and P. lucknowense, have been reported in the Indian subcontinent from shrimp, and their adults were obtained experimentally by feeding on Heteropneustes fossilis (Rai 1964; Pandey 1970). The metacercaria and adult of P. srivastava were found in the same locality and recovered from naturally infected hosts. The development of adult P. srivastava in fish is attributed to carnivorous nature of the fish host, H. fossilis, and the availability of insects and crustacean preys in its vicinity.
The comparative morphology of the present metacercaria showed a close resemblance to the adult of P. srivastava Rai 1964, including the ratio of suckers, the arrangement of testes, bipartite vesicula seminalis, and paired vitelline masses, but they differ from P. parorchium which has symmetrical testes, less crenulated margin, and deeply lobed vitelline follicles.
The morphological studies were supported by molecular findings as well. Phyllodistomum over the last decade, particularly for 28S rRNA, ITS 1, and CoxI sequences. Over the last decade, the increasing availability of genetic resources has greatly enhanced our understanding of the diversity and phylogenetic relationships of gorgoderid trematodes (Razo-Mendivil et al. 2013; de León et al. 2015a; de León et al. 2015b; Pinacho-Pinacho et al. 2021).
Regarding the comparison of base pair (bp) differences for different regions, our results align with previous studies. Razo-Mendivil et al. (2013) reported no intra-specific variation in isolates of P. inecoli and P. brevicecum for 28S and ITS1, whereas inter-specific variation ranged from 3.2 to 4.4% for 28S r DNA and from 8.3 to 14.7% for ITS1. In contrast, Petkevičiūtė et al. (2015) observed 0.3% intra-specific variation for P. folium and 0.5% for P. macrocotyle in 28S sequences. European species of Phyllodistomum, on the other hand, exhibited high inter-specific variation (8.5% to 14%). In another study by Petkevičiūtė et al. (2015), no intra-specific variation was found in 28S sequences of different isolates of European species P. folium.
Similarly, de León et al. (2015b) identified genetic variation in the 28S rRNA gene between P. spinopapillatum and P. inecoli, ranging from 0.8 to 1.0% and between P. lacustri and P. staffordi ranging from 3.8 to 4.0%. Petkevičiūtė et al. (2020) identified genetic variation in the 28S rRNA gene between P. kupermani and P. folium, ranging from 1.1 to 1.3%, whereas 1.2 to 1.4% for ITS2; while it was 1.8% for P. umblae for 28S rRNA and 1.2% for ITS2. Pinacho-Pinacho et al. (2021) identified interspecific of CoxI sequence between P. simoni and P. inecoli ranging from 5.1 to 6.4%, while 6.5–10.7% for P. spinopapillatum.
In general, the intra-specific variation within the ITS region and mt CoxI is generally low, but consistent inter-specific differences exist (Nolan and Cribb 2005; Pinacho-Pinacho et al. 2021). To obtain a clear understanding of exact number of Phyllodistomum species in India, further studies are necessary. These studies should encompass both morphological and molecular analyses of metacercariae and adults, which will significantly contribute to the phylogenetic revision of gorgoderids. The combination of these approaches will provide valuable insights into the taxonomy and evolutionary relationships of these trematode species. Certain parasites with indirect life cycles possess the capability to manipulate the behaviour of their hosts (Freire et al., 2022), thereby aiding in their transmission to definitive hosts. Given the discovery of Phyllodistomum metacercariae in shrimp and adults in fish during the current study, an interesting area for future research would involve exploring the potential impact of Phyllodistomum infection on shrimp behaviour. Certain digenean parasites, including Clinostomum and Paramphistomum, are recognized for their migration through gastrointestinal tissues before reaching adulthood in the host's stomach (Rolfe et al. 1994, Shamsi et al. 2013). This process leads to pathogenic effects in definitive hosts. Hence, an additional prospective area for future research lies in examining the health consequences and pathogenicity induced by these parasites in their fish hosts.
Data availability
Sequences are publicly available in GenBank.
References
Ahmad R, Singh V, Maurya AK (1999) On a new species of the genus Phyllodistomum Braun 1899 from the stomach of a fresh water fish Ompok bimaculatus (Bloch) at Varanasi. Adv Biosci 18:45–52
Aghlmandi F, Habibi F, Afraii MA, Abdoli A, Shamsi S (2018) Infection with metacercaria of Clinostomum complanatum (Trematoda: Clinostomidae) in freshwater fishes from Southern Caspian Sea Basin. Revue de Médecine Vétérinaire 7:147–151
Agrawal V (1966) Studies on some trematode parasites of fresh water fishes from Lucknow. Annales de Parasitologie Humaine et Comparee 41(3):217–231
Barton DP, Zhu X, Nuhoglu A, Pearce L, McLellan M, Shamsi S (2022) Parasites of selected freshwater snails in the Eastern Murray Darling Basin, Australia. Int J Environ Res Public Health 19(12):7236
Bhadauria S, Dandotia MR (1988) Studies on the digenetic trematodes of freshwater fishes with special reference to Gwalior region. Part IV. Description of ten new and six already known forms belonging to eight genera of trematodes. Rivista di Parassitologia 47(3):353–397
Blair D, Campos A, Cummings MP, Laclette JP (1996) Evolutionary biology of parasitic Platyhelminths: the role of molecular phylogenetics. Parasitol Today 12(2):66–71
Bowles J, Hope M, Tiu WU, Liu X, McManus DP (1993) Nuclear and mitochondrial genetic markers highly conserved between Chinese and Philippine Schistosoma japonicum. Acta Trop 55(4):217–229. https://doi.org/10.1016/0001-706X(93)90079-Q
Campbell R (2008) Family Gorgoderidae Looss, 1899 Keys to the Trematoda, volume 3. CABI Wallingford UK, pp 191–213
Choudhary K, Verma AK, Swaroop S, Agrawal N (2015) A review on the molecular characterization of digenean parasites using molecular markers with special reference to ITS region. Helminthologia 52(3):167–187. https://doi.org/10.1515/helmin-2015-0031
Choudhary K, Ray S, Shamsi S, Agrawal N (2022) Characterization of Clinostomum (Digenea: Clinostomidae) spp. in India. Parasitol Res 121(11):3083–3089. https://doi.org/10.1007/s00436-022-07644-y
Cribb TH (1987) Studies on gorgoderid digeneans from Australian and Asian fresh-water fishes. J Nat Hist 21(5):1129–1153. https://doi.org/10.1080/00222938700770711
Cutmore SC, Miller TL, Curran SS, Bennett MB, Cribb TH (2013) Phylogenetic relationships of the Gorgoderidae (Platyhelminthes: Trematoda), including the proposal of a new subfamily (Degeneriinae n. subfam.). Parasitol Res 112:3063–3074
Dayal J (1949) Trematode parasites of Indian fishes, part ii. Indian Journal of Helminthology 1:93–120
de León GP-P, Martinez-Aquino A, Mendoza-Garfias B (2015a) Two new species of Phyllodistomum Braun, 1899 (Digenea: Gorgoderidae), from freshwater fishes (Cyprinodontiformes: Goodeidae: Goodeinae) in central Mexico: an integrative taxonomy approach using morphology, ultrastructure and molecular phylogenetics. Zootaxa 4013(1):087–099
de León GP-P, Pinacho-Pinacho CD, Mendoza-Garfias B, García-Varela M (2015b) Phyllodistomum spinopapillatum sp. nov. (Digenea: Gorgoderidae), from the Oaxaca killifish Profundulus balsanus (Osteichthyes: Profundulidae) in Mexico, with new host and locality records of P. inecoli: morphology, ultrastructure and molecular evidence. Acta Parasitol 60:298–307
Freire R, Rogers L, Creece D, Shamsi S (2022) Neophobic behavioural responses of parasitised fish to a potential predator and baited hook. Appl Anim Behav Sci 105722
Froese R, Pauly D (2019) FishBase. World Wide Web electronic publication. www.fishbase.org, version (02/2019)
Georgieva S, Soldánová M, Pérez-del-Olmo A, Dangel DR, Sitko J, Sures B, Kostadinova A (2013) Molecular prospecting for European Diplostomum (Digenea: Diplostomidae) reveals cryptic diversity. Int J Parasitol 43(1):57–72
Goswami LM, Prasad PK, Tandon V, Chatterjee A (2009) Molecular characterization of Gastrodiscoides hominis (Platyhelminthes: Trematoda: Digenea) inferred from ITS rDNA sequence analysis. Parasitol Res 104(6):1485–1490. https://doi.org/10.1007/s00436-009-1354-8
Gupta SP (1951) On a new trematode, Phyllodistomum singhiai n. sp. of the family Gorgoderidae Looss, 1899 from the intestine of a fresh-water fish, Mastacembelus armatus (Lacep.). Indian Journal of Helminthology 3(1):21–28
Gupta SP (1955) Trematode parasites of fresh-water fishes. Indian Journal of Helminthology 5(1 1953):1–80
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hust J, Frydenberg J, Sauriau P-G, Le Gall P, Mouritsen K, Jensen KT (2004) Use of ITS rDNA for discriminating of larval stages of two microphallid (Digenea) species using Hydrobia ulvae (Pennant, 1777) and Corophium volutator (Pallas, 1766) as intermediate hosts. Parasitol Res 93:304–310
Huston DC, Cutmore SC, Cribb TH (2018) Isorchis cannoni n. sp (Digenea: Atractotrematidae) from Great Barrier Reef rabbitfishes and the molecular elucidation of its life cycle. J Helminthol 92(5):604–611. https://doi.org/10.1017/s0022149x17000906
Jaiswal G (1957) Studies on the trematode parasites of fishes and birds found in Hyderabad State. Part I-IV. Zool Jahrb Abt Syst Geogr Biol 85(1/2):1–72
Kakaji VL (1969) Studies on helminth parasites of Indian fishes. Part 3. Some trematode parasites of fresh water fishes of Uttar Pradesh. Indian Journal of Helminthology 21:49–80
Kaw BL (1950) Studies in Helminthology: Helminth parasites of Kashmir. Pt. I. Trematoda. Indian Journal of Helminthology 2:67–126
Littlewood D (2008) Platyhelminth systematics and the emergence of new characters. Parasite 15(3):333–341
Mendoza-Garfias B, de León GP-P (2005) Phyllodistomum centropomi sp. n. (Digenea: Gorgoderidae), a parasite of the fat snook, Centropomus parallelus (Osteichthyes: Centropomidae), in the Papaloapan River at Tlacotalpan, Veracruz State, Mexico. Zootaxa 1056(1):43–51
Mollaret I, Lim LHS, Justine J-L (2000) Phylogenetic position of the monogeneans Sundanonchus, Thaparocleidus, and Cichlidogyrus inferred from 28S rDNA sequences. Int J Parasitol 30(5):659–662. https://doi.org/10.1016/S0020-7519(00)00039-4
Motwani MP, Srivastava CB (1961) On two phyllodistomes from the urinary bladder of siluroid fishes (Trematoda: Gorgoderidae). Indian Journal of Helminthology 13:93–99
Mwita C, Nkwengulila G (2010) Phylogenetic relationships of the metazoan parasites of the clariid fishes of Lake Victoria inferred from partial 18S rDNA sequences. Tanzania Journal of. Science 36
Nakao M (2015) Phyllodistomum kanae sp. nov. (Trematoda: Gorgoderidae), a bladder fluke from the Ezo salamander Hynobius retardatus. Parasitol Int 64(5):314–318
Naz SS, Siddique SF (2012) Two new Digenetic trematodes Phyllodistomum parichhaii and Phyllodistomum pahujii (Family: Gorgoderidae Looss, 1901) from fresh water fish Xenentodon cancila (Ham.) of different water bodies, district Jhansi Bundelkhand region. Int J Innov Bio-Sci 2:229–231
Nolan MJ, Cribb TH (2005) The use and implications of ribosomal DNA sequencing for the discrimination of digenean species. Adv Parasitol 60:101–163
Pande PN, Dwivedi R (1983) On a new species of piscine trematode Phyllodistomum cephaloglandulata, n. sp. Read Zool 2:22–23
Pandey K (1970) On a new phyllodistome metacercaria (Phyllodistomum lucknowensis) and its adult. Indian J Zootomy 11:22–35
Pandey K, Agrawal N (2013) Metacercarial fauna of India. Zoological Survey of India Kolkatapages
Pandey KC, Agrawal N (2018) Trematode fauna of freshwater fish of India, vol 393, pp 1–388
Peribáñez MA, Ordovás L, Benito J, Benejam L, Gracia MJ, Rodellar C (2011) Prevalence and sequence comparison of Phyllodistomum folium from zebra mussel and from freshwater fish in the Ebro River. Parasitol Int 60(1):59–63
Petkevičiūtė R, Stunžėnas V, Stanevičiūtė G, Zhokhov AE (2015) European Phyllodistomum (Digenea, Gorgoderidae) and phylogenetic affinities of Cercaria duplicata based on rDNA and karyotypes. Zool Scr 44(2):191–202
Petkevičiūtė R, Zhokhov AE, Stunžėnas V, Poddubnaya LG, Stanevičiūtė G (2020) Phyllodistomum kupermani n. sp. from the European perch, Perca fluviatilis L. (Perciformes: Percidae), and redescription of Phyllodistomum macrocotyle (Lühe, 1909) with notes on the species diversity and host specificity in the European Phyllodistomum spp. (Trematoda: Gorgoderidae). Parasit Vectors 13(1):561. https://doi.org/10.1186/s13071-020-04434-2
Pinacho-Pinacho CD, Sereno-Uribe AL, Hernández-Orts JS, García-Varela M, de León GP-P (2021) Integrative taxonomy reveals an even greater diversity within the speciose genus Phyllodistomum (Platyhelminthes: Trematoda: Gorgoderidae), parasitic in the urinary bladder of Middle American freshwater fishes, with descriptions of five new species. Invertebr Syst 35(7):754–775
Prasad PK, Tandon V, Biswal DK, Goswami LM, Chatterjee A (2008) Molecular identification of the Indian liver fluke, Fasciola (Trematoda: Fasciolidae) based on the ribosomal internal transcribed spacer regions. Parasitol Res 103(6):1247–1255. https://doi.org/10.1007/s00436-008-1121-2
Rai S (1964) Observations on the life-history of Phyllodistomum srivastavai sp. nov. (Trematoda: Gorgoderidae). Parasitology 54(1):43–51
Razo-Mendivil U, Perez-Ponce de Leon G, Rubio-Godoy M (2013) Integrative taxonomy identifies a new species of Phyllodistomum (Digenea: Gorgoderidae) from the twospot livebearer, Heterandria bimaculata (Teleostei: Poeciliidae), in Central Veracruz, Mexico. Parasitol Res 112:4137–4150
Rochat EC, Blasco-Costa I, Scholz T, Unmack PJ (2020) High diversity of metazoan parasites in carp gudgeons (Eleotridae: Hypseleotris spp.) from Eastern Australia. J Helminthol 94:e146. https://doi.org/10.1017/S0022149X20000280
Rolfe PF, Boray JC, Collins GH (1994) Pathology of infection with Paramphistomum ichikawai in sheep. Int J Parasitol 24(7):995–1004
Sarwat M (2011) New species of the genus Phyllodistomum (Braun, 1899)(Digenea: Gorgoderidae, Looss, 1901) from freshwater fish Mastacembelus armatus Aurangabad (MS) India. Recent Res Sci Technol 3(8):11–13
Sen JM (2014) On a new species of Phyllodistomum Braun, 1899 (Digenea: Gorgoderidae), a parasite of fresh water fish, Channa punctatus (Bl.) from Betwa River, Bundelkhand region Jhansi, UP, India. Current World. Environment 9(1):207
Shamsi S, Banfield A, Francis N, Barton DP, McLellan M (2023) Occurrence of digenean parasites in freshwater snails in the Murrumbidgee catchment area. Australia. Food and Waterborne Parasitology e00202
Shamsi S, Barton DP, Day S, Masiga J, Zhu X, McLellan M (2021a) Characterization of Clinostomum sp. (Trematoda: Clinostomidae) infecting cormorants in south-eastern Australia. Parasitol Res 120:2793–2803. https://doi.org/10.1007/s00436-021-07246-0
Shamsi S, Day S, Zhu X, McLellan M, Barton DP, Dang M, Nowak BF (2021b) Wild fish as reservoirs of parasites on Australian Murray Cod farms. Aquaculture:736584. https://doi.org/10.1016/j.aquaculture.2021.736584
Shamsi S, Halajian A, Tavakol S, Mortazavi P, Boulton J (2013) Pathogenicity ofClinostomum complanatum (Digenea: Clinostomidae) in piscivorous birds. Res Vet Sci 95(2):537–539
Shomorendra M, Jha AN (2006) On a new trematode parasite Phyllodistomum guptai n.sp. of the family Gorgoderidae Looss 1901, from body cavity of freshwater fishes. Uttar Pradesh Journal of Zoology 26(3):339–342
Singh SP, Sinha DP (1975) Two new trematodes from fresh water fishes of Bihar. In: Tiwari KK, Srivastava C (eds) Dr. B.S. Chauhan commemoration volume. The Zoological Society of India, Vani Vihar, Orissa, India, pp 411–417
Srivastava HD (1938) A new gorgoderid trematode from the urinary bladder of an Indian migratory fish, Belone strongylura. Indian Journal of Veterinary Science 8:391–393
Stunžėnas V, Petkevičiūtė R, Poddubnaya LG, Stanevičiūtė G, Zhokhov AE (2017) Host specificity, molecular phylogeny and morphological differences of Phyllodistomum pseudofolium Nybelin, 1926 and Phyllodistomum angulatum Linstow, 1907 (Trematoda: Gorgoderidae) with notes on Eurasian ruffe as final host for Phyllodistomum spp. Parasit Vectors 10(1):1–15
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Tkach V, Pawlowski J, Mariaux J (2000) Phylogenetic analysis of the suborder Plagiorchiata (Platyhelminthes, Digenea) based on partial lsrDNA sequences. Int J Parasitol 30(1):83–93
Urabe M, Ishibashi R, Uehara K (2015) The life cycle and molecular phylogeny of a gorgoderid trematode recorded from the mussel Nodularia douglasiae in the Yodo River, Japan. Parasitol Int 64(1):26–32
Yamaguti S (1934) Studies on the helminth fauna of Japan. Part 2, Trematodes of fishes, I. Japanese. J Zool 5(3):249–541
Yamaguti S (1958) Systema helminthum, vol 1. The digenetic trematodes of vertebrates. Interscience Publications, New York, pp 1–1575
Acknowledgements
Financial assistance to K. Choudhary under Rajiv Gandhi National Fellowship of UGC: F1-17.1/2013-14/ RGNF-SC-UTT-51511/ (SA-III) are acknowledged. Facilities developed under UGC-SAP (DRS-I & II), DST-PURSE, UGC-BSR (NA F-4-10/2010 BSR), (NA EMERITUS-2017-18-GEN-9293/SA-III) Department of Zoology, University of Lucknow, Lucknow, were also utilized for the present work. Authors are grateful to Mt Craig Poynter from CSU for creating the map of the study area.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions
Author information
Authors and Affiliations
Contributions
Kirti Choudhary: Parasite collections, laboratory work, microscopy, phylogenetic trees. Shailendra Ray: Parasite collections, laboratory work, microscopy, phylogenetic trees. Nirupama Agrawal: Principal supervisor; study design; parasite identification; manuscript writing. Shokoofeh Shamsi: Data analyses, supervision, manuscript writing
Corresponding author
Ethics declarations
Ethical approval
This work was conducted under animal ethic approval 20/I/2023/IAEC/LU.
Consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Handling Editor: Una Ryan
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Choudhary, K., Ray, S., Agrawal, N. et al. Genetic characterization and phylogenetic relationships of Phyllodistomum parasites in Indian subcontinent: insights from freshwater fish and shrimp hosts. Parasitol Res 122, 2301–2315 (2023). https://doi.org/10.1007/s00436-023-07930-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-023-07930-3