Abstract
Dynamic networks are valuable tools to depict and investigate the concurrent and temporal interdependencies of various variables across time. Although several software packages for computing and drawing dynamic networks have been developed, software that allows investigating the pairwise associations between a set of binary intensive longitudinal variables is still missing. To fill this gap, this paper introduces an R package that yields contingency measure-based networks (ConNEcT). ConNEcT implements different contingency measures: proportion of agreement, corrected and classic Jaccard index, phi correlation coefficient, Cohen’s kappa, odds ratio, and log odds ratio. Moreover, users can easily add alternative measures, if needed. Importantly, ConNEcT also allows conducting non-parametric significance tests on the obtained contingency values that correct for the inherent serial dependence in the time series, through a permutation approach or model-based simulation. In this paper, we provide an overview of all available ConNEcT features and showcase their usage. Addressing a major question that users are likely to have, we also discuss similarities and differences of the included contingency measures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last decennium, a surge of network methods washed up on the shores of the behavioral sciences. Networks offer valuable tools to depict and investigate the complex interdependencies of various variables. The variables constitute the nodes of the obtained networks and the strength of the pairwise or conditional (upon other variables) associations between the variables are represented through the edges that connect the nodes. Network methods have been applied to a wide range of problems, including affective dynamics (e.g., Bodner et al., 2018; Bringmann et al., 2016), attitudes (e.g., Dalege et al., 2016), beliefs (e.g., Brandt et al., 2019), psychopathology (Borsboom, 2008, 2017; Borsboom & Cramer, 2013; Cramer et al., 2010; Fried et al., 2017), and parent-child interactions (Bodner et al., 2018, 2019; Van keer et al., 2019).
Network methods come in variations. First, the underlying data can be cross-sectional (i.e., many individuals are measured once) or intensive longitudinal (i.e., one or more individuals are measured frequently). Psychopathological networks, for example, were initially built based on cross-sectional data, facilitating insight into how symptoms relate across individuals (e.g., Boschloo et al., 2015; Cramer et al., 2016; Isvoranu et al., 2016). Complementing these cross-sectional networks with dynamic networks, built on intensive longitudinal data, sheds additional light on the within-subject relations between variables over time (e.g., Bringmann et al., 2013; Bulteel, Tuerlinckx, et al., 2018b; Epskamp, Waldorp, et al., 2018b; Hamaker et al., 2018). Second, the underlying data can be continuous, ordinal, categorical and binary (e.g., absence or presence of a behavior), or combinations thereof (mixed data). While most methods have been developed for continuous data (e.g., Epskamp, Waldorp, et al., 2018b), attention has also been paid to binary data (e.g., van Borkulo et al., 2014) and mixed data (Haslbeck & Waldorp, 2020). Third, while most network methods focus on statistical model-based conditional variable associations, quantified through partial correlations (e.g., Epskamp, Borsboom, & Fried, 2018a; Lafit et al., 2019) or regression weights (e.g., Bulteel et al., 2016a), some approaches investigate and explore simple bivariate (or pairwise) associations, without making model-based assumptions about how these connections come about (e.g., Bodner et al., in press). An appealing feature of studying bivariate relations is that they are not affected by the composition of the variable set, whereas conditional associations may change when a variable is added or excluded. On the other hand, model-based network approaches are obviously attractive in that they may offer deeper insight into the mechanisms behind the observed associations. In the end, as holds for statistical analyses in general, the research question at hand should determine whether to focus on conditional or unconditional associations.
Scrutinizing the available software with the above three distinctions in mind, the development focused on conditional association-based approaches for continuous cross-sectional data (e.g., Epskamp & Fried, 2018). Software for the analysis of binary data (e.g., mgm, Haslbeck & Waldorp, 2020; IsingFit, van Borkulo et al., 2014) and intensive longitudinal data (e.g., mlvar, Epskamp, Waldorp, et al., 2018b) has also been proposed. However, software for networks based on pairwise associations of binary intensive longitudinal variables is still missing, although such approaches have led to meaningful insights when investigating micro-coded parent-child interactions (Bodner et al., 2018, 2019; Van keer et al., 2019) and longitudinal depression symptom reports (Bodner et al., in press). Therefore, we propose an R-package for building such contingency measure-based networks, which we called ConNEcT (Bodner & Ceulemans, 2021). The ConNEcT package includes seven contingency measures: proportion of agreement, the classic and the corrected Jaccard index, phi correlation coefficient, Cohen’s kappa, odds ratio, and log odds ratio. Other contingency measures can be easily added, as we will demonstrate. The package can be used to investigate concurrent associations (e.g., the association between two behaviors X and Y at the same moment) as well as temporal sequences (e.g., is the presence of behavior X at time point t associated with the presence of behavior Y at time point t+δ). The ConNEcT software also provides a tailor-made significance testing framework (Bodner et al., 2021). Finally, the package allows the visualization of the results in network figures.
The paper is organized into four modules, focusing on data requirements and exploration, contingency measure selection and computation, significance testing, and network visualization, respectively. Each module first gives a theoretical introduction to the topic, where we also delve deeper into some so far unanswered questions, such as the similarities and differences of the included contingency measures. Next, we explain how to apply the ConNEcT R-Package using illustrative examples. Figure 1 gives a visual overview of the four modules, making use of intensive longitudinal depression symptom data from a patient included in the study by Hosenfeld et al. (2015), which was also analyzed by Bodner et al. (in press).
Overview of the four ConNEcT modules using the data of weekly reported depression symptoms. This example patient (Hosenfeld et al., 2015) reported the presence or absence of eight depression symptoms (Core symptoms, lack of energy, eating problems, sleeping problems, psychomotor problems, feelings of guilt, cognitive problems, and preoccupation with death) for 145 weeks. a Line plot of the depression symptoms over weeks, where elevated line segments indicate presence and segments that coincide with the reference line indicate absence. b Heatmap of the strength of the pairwise classic Jaccard values, quantifying concurrent contingency. c Histogram of the sampling distribution of the classic Jaccard value; the solid line indicates the observed Jaccard value and the dashed line the 95th percentile. d Network of the significant (α=0.05) contingencies; the node size reflects the relative frequency of the variables, while the saturation and width of the undirected edges represent the strength of the concurrent contingency.
Module 1: Data requirements and exploration
ConNEcT has been developed to investigate bivariate contingencies in binary time series data. Such data show up in different forms in the behavioral sciences. To acknowledge this variety, we will make use of three data examples to illustrate the possibilities. The datasets are also included in the package.
-
(1)
Symptom Data. The depression symptom data (see Fig. 1) stem from a patient in the study of Hosenfeld et al. (2015), also used in Bodner et al. (in press). The patient reported each week on the presence or absence of eight depression symptoms (core symptoms, lack of energy, eating problems, slee** problems, psychomotor problems, feelings of guilt, cognitive problems, and preoccupation with death) for 145 weeks. This data set is used in the introduction and in Modules 1 and 3.
-
(2)
Family Data. These data, collected by Sheeber et al. (2012) and re-analyzed by Bodner et al. (2018), stem from a family interaction between two parents and their adolescent son or daughter during a nine-minute problem-solving interaction. The presence and absence of expressions of ‘anger’, ‘dysphoric’ feelings, and ‘happiness’ were coded for each family member in an event-based way (i.e., noting when a certain behavior starts and when it stops). The codes were subsequently restructured into second-to-second interval data, resulting in a 540 seconds by nine variables binary data set. This data set is used in Modules 2 and 4.
-
(3)
Attachment Data. In an attachment study (Bodner et al., 2019; Dujardin et al., 2016), a mother and her child (aged 8 to 12 years) were videotaped while working on a three-minute stressful puzzle task. The interaction was coded in two-second intervals for the presence and absence of positive, negative, or task-related behavior. The data set contains seven variables (‘Mother positive’, ‘Mother negative’, ‘Mother working alone’, ‘Mother and child working together’, ‘Child positive’, ‘Child negative’ and ‘Child working alone’) and 90 time points. This data set will be used in Module 3 and 4.
Theory
Number of variables and time points
Before starting the analysis, we recommend careful consideration of which variables should be included. Though the values for pairwise contingency measures are not influenced by the total set of variables (in contrast to multivariate models in which parameter estimates can change when additional variables are modeled), including a high number of variables can have consequences for the interpretability of the networks as it may imply that the network may become a complex, hard to interpret tangle of links and nodes.
The optimal number of time points depends on two different types of considerations. First, the obtained number of time points is determined by the length of the covered time period and the frequency of the measurements, with longer time periods and higher frequency leading to longer time series. In case one is interested in rare (e.g., physical aggression) and short-lived behaviors (e.g., eye-movements), the overall time period should be long and the measuring frequent, to end up with a sufficient amount of time points at which the behavior is shown. For longer-lasting and more frequently occurring behaviors shorter and coarser time series may do. The second type of consideration pertains to statistical power. Longer time series will increase the available amount of information and decrease the estimation uncertainty of the contingency strength, and thus increase the power of significance tests (see Module 3).
Relative frequency and serial dependence
During data exploration, two characteristics of time series data are especially important to consider: the relative frequency (i.e., the proportion of ‘1’s; see above and Module 3) and the serial dependence (or auto-dependence) of each variable. Variables with very high or very low relative frequencies may not be very informative, since the absolute values of many contingency measures become hard to interpret in case of extreme relative frequencies (see Module 3). Additionally, a simulation study by Brusco et al. (2021) indicates that some contingency measures lead to comparable contingency values for certain relative frequencies but not for others, again suggesting that the relative frequency of the variables under study might be important to consider when deciding which contingency measure to use.
Serial dependence refers to the tendency of behaviors to be present for more than one time point. We can quantify it by calculating conditional probabilities and comparing them to each other or to the relative frequencies. Specifically, if the probability that a ‘1’ is observed given that a ‘1’ has been observed the time point before, p(Xt=1|Xt-1=1) or p1 ∣ 1 for ease of notation, differs from the probability that a ‘1’ is shown given a zero at the previous time point, p(Xt=1|Xt-1=0) or p1 ∣ 0, or from the relative frequency of ‘1’s, p(Xt=1) or p1, this suggests that serial dependence is present. Accounting for such serial dependence is important, to avoid false positives during significance testing (Bodner et al., 2021).
Tutorial
The ConNEcT package offers some data exploration features. Users can visualize the course of the raw data over time, as well as calculate and visualize relative frequency and auto-dependence.
Basics of the conData function
The input for the conData function is the raw data, structured in a time-points-by-variable matrix. Missing values need to be retained because certain operations (e.g., lagging the data; see Module 2) might lead to erroneous contingencies when missing values have already been removed. The function removes columns that contain non-binary values (e.g., identity number or time interval counting) and calculates the relative frequency and conditional probabilities p1 ∣ 1and p1 ∣ 0 of each variable (see 3.2.3). The output is a conData object, containing data: the raw data, after removing continuous and non-binary variables, and probs, a table containing the relative frequencies and conditional probabilities of all variables. The labels of the variables are stored in varNames.
Relative frequency and serial dependence
To examine the relative frequency and the auto-dependence of each variable, we can examine the contents of the probs field of the conData object (Table 1). The symptom Death, for example, is shown 65% of the time (i.e., 94 of the 145 time points). Moreover, this symptom is characterized by high auto-dependence, since almost every 1-score is followed by another 1-score (i.e., 90 out of the 94 times, resulting in a p1 ∣ 1 of .96 and p0 ∣ 1of .04), while a 1-score rarely follows a 0-score (4 out of the 51 times, resulting in p1 ∣ 0 of .08 and p0 ∣ 0 of .92).

Visualizations
The package provides different visualizations of the raw data that might reveal interesting characteristics, using the plot function. We will illustrate the function with the Symptom Data (see Fig. 1). First, the conData function is applied to the data and the results are saved in a conData object that we call Sdata. The plot.conData function can simply be called plot(Sdata). The plottype of the output can be specified as ‘interval’, ‘line’, or ‘both’. Figure 2 shows the plot type ‘interval’, displaying a vertical tick each time a symptom was reported. The time intervals are indicated on the x-axis, while the y-axis represents the different symptoms. Alternatively, the plot type ‘line’ (see Fig. 1a) shows the presence of a variable in terms of the height of the line, where the line is higher than (respectively, coinciding with) the grey dotted auxiliary line indicates that the symptom is present (respectively, absent). It is also possible to choose ‘both’ to get a line plot with ticks at all intervals.

The relative frequencies can be visualized employing the barplot functionFootnote 1. This function has two different plot types, plotting either only the relative frequency (plottype=‘RelFreq’; see Fig. 3) or all three probabilities p1, p1 ∣ 1, and p1 ∣ 0 (plottype=‘All’; see supplementary material https://osf.io/p5ywg/).

Module 2: Contingency measure selection and computation
Theory
To quantify the strength of the bivariate association between each pair of variables (X, Y), contingency values are computed. In the literature, a myriad of contingency measures has been proposed. They are often distinguished along two lines (e.g., Brusco et al., 2021; Warrens, 2008a): First, they differ in whether contingencies are assessed while accounting for the co-occurrence of zeros or not. Second, while some measures do not account for the amount of agreement that can be expected based on the relative frequencies of the variables, others compensate for this expected amount of agreement and are designed to have a zero value if the variables can be considered statistically independent. To represent these distinctionsFootnote 2, the following popular contingency measures were included in the ConNEcT-package: proportion of agreement (co-occurrence of zeros included, no correction), classic (co-occurrence of zeros ignored, no correction), and corrected Jaccard index (co-occurrence of zeros ignored, corrected), Cohen’s kappa (co-occurrence of zeros included, corrected), and the phi correlation coefficient (co-occurrence of zeros included, corrected). We also included the (log) odds ratio, because of its relationship to logistic regression, which often underlies model-based network approaches for binary data (e.g., van Borkulo et al., 2014). Interestingly, the chosen coefficients also represent the three biggest clusters of contingency measures discussed by Brusco et al. (2021).
In what follows, we will first introduce the different contingency measures. Second, we will investigate how the measures relate to each other. We will especially focus on the domain in which each measure is defined (the findings are related to the definitions of the measures in the Appendix) and the correlations between the measures. We illustrate these relations by applying the different measures to empirical data. Third, we will explain how contingency measures can also be used to investigate the temporal relations between variables.
Introduction of the contingency measures
Notation
Table 2 shows the crosstabulation of a variable pair (X, Y), where the values ‘1’ and ‘0’ indicate the two possible values of the variables, indicating, for example, whether a certain behavior is shown (‘1’) or not (‘0’) or whether a symptom is reported as present (‘1’) or as absent (‘0’). In this table, a denotes the proportion of time points at which both variables equal ‘1’, b the proportion that only variable X equals ‘1’, c the proportion that only variable Y equals ‘1’, and d the proportion that both variables equal ‘0’. We also use the relative frequencies \({p}_1^X\) and \({p}_1^Y\) that were introduced in Module 1, and their complements \({p}_0^X\) and \({p}_0^Y\). Note that these relative frequencies equal sums of a, b, c, and d. For example, \({p}_1^X=a+b\), \({p}_1^Y=a+c\).
Proportion of agreement
The proportion of agreement PA quantifies the observed agreement, that is the proportion of time points at which the values of both variables equal either ‘1’ or ‘0’:
This measure ranges from 0 to 1, with 1 indicating perfect agreement and 0 perfect non-agreement (every 1 in one time series pairs with a 0 in the other and vice versa). The proportion of agreement attaches equal importance to the co-occurrence of the values ‘1’ and of the values ‘0’. This for instance makes sense when analyzing dichotomous data on failed and passed tests (see Brusco et al., 2021) as including both double failures and double passes allows to shed light on aptitudes and learning deficits. This contingency measure is always defined.
Cohen’s kappa
Since the obtained proportion of agreement is affected by the relative frequencies of the variables (Bodner et al., 2021), Cohen’s kappa (Cohen, 1960) refines it by correcting for chance agreement
where PE is computed as follows:
Since PA = a + d, see Eq. (1), and \({p}_1^X=a+b\), etc. we can simplify to (Warrens, 2008b):
Cohen’s kappa ranges from -1 to 1 with a value of 0 indicating that the two variables are statistically independent. From Eq. (4) we derive that this situation occurs if ad equals bc. Moreover, kappa is not defined (n.d.) in the cases where both time series contain only ‘0’s or ’1’s, as in such cases not only the numerator but also the denominator of (4) reduces to zero.
Classic Jaccard index
The Jaccard index was introduced by Jaccard (1901, 1912), to measure the ecological similarity of different geographical regions, based on the co-occurrence of specific species. It is calculated as:
The Jaccard index thus equals the proportion of time points at which both variables equal ‘1’ over the proportion of time points at which at least one of them is shown. This means that the Jaccard index only depends on the number of time points, in which the values of both variables equal ‘1’, but ignores those where both equal ‘0’.
This measure may, therefore, be useful for behavioral science questions, for which the co-absence of symptoms or behaviors is of less importance than their co-occurrence (Bodner et al., in press; Brusco et al., 2019). For example, Main et al. (2016, p. 915) argue that they ‘do not wish to treat shared absence of a target emotion in two people as a kind of synchrony of that emotion.’ The co-absence of emotions and behaviors in micro-coded interaction data is indeed often a somewhat artificial result of assigning each coded event to a single coding category only, implying that the presence of one variable, automatically implies the absence of other variables (e.g., SPAFF, Coan & Gottman, 2007; LIFE, Hops et al., 1995). The Jaccard index ranges from 0 to 1 and is not defined if both time series have a relative frequency of 0.
Corrected Jaccard index
Like the proportion of agreement, the classic Jaccard measure (JA) does not correct for chance agreement. Therefore, Bodner et al. (2019) developed a corrected Jaccard index (JCorr), in which the classic Jaccard index is compared to an expected value (JE), computed using the principles outlined in Albatineh and Niewiadomska-Bugaj (2011):
J E expresses the Jaccard value that we would expect if X and Y do not systematically co-occur. It only depends on the relative frequencies of X and Y:
Whereas a corrected Jaccard value of 0 implies that X and Y do not co-occur more than expected by chance, a negative value indicates that X and Y co-occur less than expected by chance. The corrected Jaccard is not defined if both time series have a relative frequency of 0 or of 1.
Odds ratio and log odds ratio
The odds ratio OR(X, Y) and the log odds ratio LOR(X, Y) are defined as
The odds ratio ranges between 0 and +Infinity, with a value of 1 (i.e., ad = bc ), indicating statistical independence between the variables. The value is not defined ( +Infinity), if b and/or c equals zero, implying that X or Y does not occur without the other (see also Bodner et al., 2021). The value is 0 when a and/or d equal zero, implying that X and Y do neither co-occur nor are they co-absent. In these cases, the value of the log odds ratio is not defined (-Infinity). The log odds ratio, therefore, ranges between -Infinity and +Infinity, with a value of zero indicating that statistical dependence cannot be assumed. Finally, both indices are also not defined if at least one of the variables has a relative frequency of 0 or 1.
Phi correlation coefficient
The phi correlation coefficient (Yule, 1912) equals the Pearson correlation coefficient computed on binary data. It can be calculated as:
The phi-correlation coefficient, therefore, has a value between -1 (perfect disagreement) and 1 (perfect agreement), with 0 indicating statistical independence. Like the odds ratio and the log odds ratio, the formula of the phi correlation coefficient features a product in the denominator. As for them, variables with a frequency of 1 or 0 imply the phi value to be undefined. In contrast to those two measures, the phi correlation coefficient, however, is never infinite.
Although the phi correlation coefficient is more often not defined than Cohen’s kappa (see Table 3 and the Appendix), some equalities between these two measures strike the eye. First, both Cohen’s kappa and the phi correlation coefficient equal 0 if ad = bc (if none of the variables has the frequency 0 or 1). Second, it can be derived that rφ(X, Y) exactly equals k(X, Y) whenever \({p}_1^X={p}_1^Y\).
Other contingency measures
The discussed contingency measures are only a subset of all possible measures (see Brusco et al., 2021; Warrens, 2008b). Other measures have been proposed, such as Yule’s Q (Bakeman et al., 1996; Bakeman & Quera, 2011) and the risk difference (Lloyd et al., 2016), the recurrence rate of cross-recurrence quantification analysis (Main et al., 2016) and Wampold’s transformed kappa (Bakeman et al., 1996; Holloway et al., 1990). Measures that were developed for interrater reliability, like Gwet’s AC (Gwet, 2014; Wongpakaran et al., 2013), Bangidwala’s B (Munoz & Bangdiwala, 1997) or Yule’s Y (Yule, 1912), or measures developed for comparing partitions, like the Rand index (Rand, 1971) or the adjusted Rand Index (Hubert & Arabie, 1985) might also be suitable. In the Tutorial part, we will demonstrate how such alternative contingency measures can be added to the package.
Impact of the differences between the seven considered contingency measures
Contingency measures have been developed in many different domains (biology, economy, interrater agreement, partitioning, etc.; Warrens, 2008b). Many of these measures are hand-tailored for one specific context and use a specific notation, which makes a direct comparison of their definitions difficult. Therefore, contingency measures are often compared by calculating their values on simulated or empirical data (Brusco et al., 2021; Todeschini et al., 2012). Likewise, we will investigate the differences between the seven contingency measures in practice by calculating their values on empirical data and comparing the resulting values. To this end, we analyzed 4860 time series pairs taken from the study from which the Family Data (see Module 1) were also taken. The data represent the interaction data of several different families. The variables have a prevalence ranging between 0 and .94 with a mean of .12.
First, we investigate the domains in which each measure is (not) defined. Table 3 summarizes the conditions that lead to not defined values, be it unspecified (n.d.) or infinite (+Inf or -Inf) in these 4860 variable pairs and their prevalence for the different contingency measures (as discussed in section 12). In the Appendix, we investigate how these findings can be explained by the definitions of the contingency measures. There, we also discuss all potential cases that lead to not-defined values also including those cases that are not shown in this data, for example, those for variables with prevalence 1 (see Appendix Table A1, A2, A3).
Second, we investigate how the different measures relate to each other. The values of the contingency measures, especially their means and standard deviations (Table 4), were quite different. As a direct comparison, therefore, was difficult, we investigated whether the rank order was comparable between measures by calculating the Spearman rank correlations across the 2491 binary variable pairs yielding defined and finite values for all contingency measure (2369 out of the 4860 variable pairs from the sample above led to undefined values for at least one contingency measure; see Table 3). The corresponding scatter plot matrix and the distributions of the contingency values can be consulted in the Appendix (Figure A1).
Table 4 shows that Cohen’s kappa, corrected Jaccard, and phi correlation coefficient have very high rank order correlations, whereas the proportion of agreement yields the most deviating ranking, followed by the classic Jaccard. Figure 4 investigates the differences between kappa, the corrected Jaccard, and phi in more detail by plotting their values as a function of the obtained kappa values. The figure reveals that the three indices coincide when kappa equals 0, but deviate elsewhere. Phi and kappa values are equal in some cases (e.g., when the relative frequencies of both variables equal each other), but in general phi yields higher absolute values. The corrected Jaccard, in contrast, yields less extreme values than kappa.
Relationship between Cohen’s kappa, corrected Jaccard and phi correlation coefficient for 3698 pairs binary time series (1162 of the 4860 pairs of the original sample yielded undefined values for at least one of the contingency measures; see Table 3). The triplets are sorted based on Cohen’s kappa values.
From concurrent to temporal relations
So far, we have only discussed concurrent bivariate relations. The temporal sequencing of variables can be used to investigate whether the presence of variable X at time point t is linked to the presence of variable Y one or more time points later, or vice versa. For instance, in coded interaction tasks the behavioral sequences between the interacting individuals are often studied (Bodner et al., 2018). Interestingly, such questions can be investigated by implementing the very same contingency measures, after appropriately lagging one of the two variables. For instance, to assess the strength of the association between X (at one time point) and Y (at the next time point), we calculate the contingency of Xt and Yt+1, implying a lag of one on Y. Per pair (X, Y), we always examine two temporal relations, one where only X is lagged and one where only Y is lagged; in both cases the same lag is used. This makes it, for example, possible to investigate whether both directions are significant or only one of them (or none).
An important question, however, is how to determine how large this lag should be. Most often this decision is taken based on previous findings or the research hypothesis. When there are no clues, however, one can obtain a tentative decision by considering different lags and evaluating how the values of the contingency measure change across different lags, through the inspection of a contingency profile. Specifically, we advise checking at which lag the contingency value becomes maximal and retaining this lag. The idea is based on a study by Main et al. (2016)Footnote 3. To illustrate this principle, we plotted two contingency profiles for the Family Data showing how the classical Jaccard index fluctuates across different lags (Fig. 5). The contingency profile for the Jaccard association between ‘father happy’ and ‘adolescent happy’ (panel a), shows that the Jaccard value is maximal at a lag of 0 (dashed line), providing evidence of a concurrent association (e.g., they probably laugh a lot together). In panel b, the Jaccard value reaches its maximum at a lag of 4, suggesting a leader-follower behavioral sequence, where the adolescent’s anger leads to an angry reaction of the mother four seconds later. Although these contingency profiles shed interesting light on the temporal dependence structure, they also indicate that finding a one-lag-fits-all-associations solution may not be very realistic. Therefore, it might make sense to apply the different indicated lags on the same data and compare the resulting networks, to shed light on which contingencies are concurrent and which are of a fast- or slow-reacting nature (Van keer et al., 2019; Wilderjans et al., 2014). Note that the described procedure to select the optimal lag is descriptive, in that no significance test is performed. Investigating how to perform such lag selection tests is an interesting direction for future research.
Tutorial
Using the Family Data, we first discuss the conMx function that is provided to compute a contingency matrix and show how to visualize it in a heatmap using corrplot (Wei & Simko, 2021). Second, we illustrate how to obtain contingency profiles. Finally, we explain how alternative contingency measures can be integrated into the package.
Basics of the conMx function
The conMx function takes the following input arguments:
-
data: Raw time series data in a time-point-by-variable matrix. Before calculating the contingency values, the non-binary variables will automatically be removed using the conData function (See Module1).
-
lag: (default lag=0, indicating co-occurrence of data) a non-negative number, indicating the intervals for which the subsequent data shall be lagged. Lag=5 means that the contingency measure is calculated between all possible pairs Xt and Yt+5 (thus also between Yt and Xt+5,).
-
conFun: The function used to calculate the contingency measure. For now, the classic Jaccard index (funClassJacc), corrected Jaccard (funCorrJacc), Cohen’s kappa (funKappa), proportion of agreement (funPropAgree), odds ratio (funOdds) and log odds ratio (funLogOdds) and the phi correlation coefficient (funPhiCC) are included.
and yields a conMx object as output, with two fields
-
value, a V by V matrix, with V indicating the number of variables. If a lag larger than 0 is used, the rows of the asymmetric matrix indicate the leading variable, and the columns the following variable. If the lag equals zero, the matrix is symmetric.
-
para, the parameters of the analyses, subdivided in the lag field (lag), the name of the contingency function (funName), and the names of the variables (varNames).
Calculating contingency matrices
Let us now take a look at the Family Data. We use a lag of one second to investigate the fast temporal sequencing of the emotional expressions, and the classic Jaccard measure to focus on the sequential co-occurrence of emotional reactions.

To visualize the contingency matrix as a heatmap, we make use of the corrplot function from the R package corrplot (Wei & Simko, 2021). The name of the contingency measure used to compute contingencies is added with a mtext to the plot after extracting it from the conMx$para$funName field.

Panel a in Fig. 6 shows the Jaccard contingency matrix. To find the contingency between ‘adolescent happy’ and ‘mother happy’ one second later we look for the J(adhappy, mohappy) value (in the last row and the seventh column) which equals .33. The other panels in Fig. 6 provide the contingency matrices for Cohen’s kappa, the corrected Jaccard, and the phi correlation coefficient Fig. 7.
Calculating and visualizing a contingency profile
To assess which lag is most suitable for investigating the temporal dependencies, we make use of contingency profiles (see Fig. 5). A contingency profile plots the contingency between two variables for different lags. We can make this plot using the conProf function, which needs as input the data (data) and the contingency measure of interest (conFun). We also need to specify the maximum lag of interest (maxlag). The output is a conProf object that consists of a value that includes (maxlag*2+1) contingency matrices for the different lags (ranging from -maxlag over zero to +maxlag), the parameters (para) containing the maximum lag (maxLags), the name of the contingency function (funName) and the names of the variables (varNames). The contingency profile is drawn using the following codeFootnote 4:

The contingency profiles are arranged like a matrix. For instance, the second contingency profile in the first row pertains to the contingency between mohappy and fahappy. Whereas the positive lags indicate that mohappy leads and fahappy follows, the negative lags represent the values for the reverse direction (i.e., fahappy followed by mohappy). Therefore, the two off-diagonal plots per variable pair are mirrored versions of one another. The profiles on the diagonal are the auto-profiles, which reach their maximum at a lag of 0.
Experts’ excursion: How to integrate another contingency measure
The functions that are already included in the package cover a wide range of possibilities. However, a specific research question (e.g., an extension to existing research) might ask for a specific contingency measure that is not yet included. The ConNEcT package allows to include other contingency measures. We will showcase this by including the calculation of Gwet’s AC1 (Gwet, 2014; Wongpakaran et al., 2013), an interrater agreement measure for binary data, that corrects for chance agreement. In the first step, the calculation of the measure should be specified, either manually or by calling a function. Here we make use of the function gwet.ac1.raw of the irrCAC-package (Gwet, 2019). The value for Gwet’s AC can be retrieved by gwet.ac1.raw(x)$est$coeff.val and is stored in the value field. Second, the function must be given a name, both in the funName field and in the title. Finally, the function should be saved in an R -file using the same name (funGwet.R).

The function needs to be loaded using source() and the dependencies installed and loaded before running any function on the newly created contingency measures function.

Module 3: Testing Significance
Theory
Many network methods prune network edges to avoid that further investigations are influenced by links of which the value is mainly an artifact of the modeling technique (e.g., Epskamp & Fried, 2018). This is often achieved through a regularization approach (e.g., Bulteel et al., 2016b; Kuismin & Sillanpää, 2017; Lafit et al., 2019). Implementing such approaches is not possible, however, when no modeling approach is used. It might nevertheless be interesting to prune a network that relies on simple bivariate relations. One alternative strategy that one might think of is to simply prune edges based on the contingency strength, using some overall threshold value. We do not recommend this strategy because the contingency strength might depend on the relative frequency (Brusco et al., 2021) and the auto-dependence of the variables (Bodner et al., 2021), as we now elucidate further. Figure 8 (first row) illustrates the relationship between the contingency measures and the relative frequency by plotting the contingency strength for two independently generated variables without auto-dependence as a function of their relative frequency. First, some contingency measures highly depend on relative frequency: we observe a direct impact of the relative frequency on the mean of the obtained values in the U-shaped relation for the proportion of agreement and an upward trend for the classic Jaccard. The mean of the contingency measures that correct for the relative frequency—kappa, corrected Jaccard, phi—remains close to zero, as desired. Second, the range and/or standard deviation might depend on the relative frequency: the observed range for the log odds ratio, for example, is much higher for the extreme relative frequencies, though the mean remains stable. Third, all contingency measures discussed here depend on serial dependence. The second to fourth rows of Fig. 8 further put the spotlight on the range of the obtained values, now focusing on auto-dependence. These rows show the distribution of the obtained values for two independent variables with a relative frequency of .5 and with an auto-dependence that arises from none (second row) over moderate (third row) to strong (fourth row). The range of the obtained values increases for higher levels of auto-dependence. This makes sense as auto-dependence may artificially install overlap, which can be mistaken for contingency. Specifically, two variables that show identical values at least once and contain longer periods of the same values within each variable, are more likely to show identical values also on adjacent time points. As a consequence, we observe broader sampling distributions, for higher serial dependence.
Dependence of contingency values on the relative frequency and serial dependence. The first row plots the contingency values as a function of relative frequencies, with the different lines representing the mean (solid line), +/- 1 sd (dashed lines), and min/max (dotted lines). The values are calculated based on simulated, independent variables X and Y (\({p}_1^{\mathrm{X}}={p}_1^{\mathrm{Y}}\)range between 0.05 and 0.95; each pair contains 500 time points) without auto-dependency (\({p}_1^{\mathrm{X}}={p}_{1\mid 1}^{\mathrm{X}}={p}_{1\mid 0}^{\mathrm{X}}={p}_1^{\mathrm{Y}}={p}_{1\mid 1}^{\mathrm{Y}}={p}_{1\mid 0}^{\mathrm{Y}}\)). Row 2-4: Distributions of the contingency measures. While the relative frequency of X and Y always equals .5, their auto-dependency varies across the rows, in that in the second row p1|1=p1|0=0.5 (light-grey), in the third row, p1|1=0.8 (middle-grey), and in the bottom row p1|1=0.95 (black). Each panel is based on 100 replicates of the pairwise independent variables X and Y (each consisting of 500 time points) on which contingency values were calculated for all 10,000 possible pairs.
Therefore, we propose to prune network edges using the non-parametric significance testing procedure introduced and extensively validated by Bodner et al. (2021). This procedure investigates whether a certain contingency value is bigger than would be expected by chance, given the relative frequency and auto-dependency of the data. In what follows we briefly summarize the main principles of the test and explain the different variants that are included in ConNEcT.
Basic principles
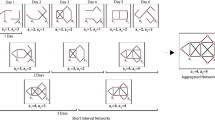
The significance test consists of five steps, depicted in Fig. 9. After collecting the time series data (step 1) and calculating the chosen contingency measure on a pair of variables (step 2), N samples or replicates of surrogate variable pairs are generated (step 3), under the null hypothesis that both variables are independent. This generation mechanism of the surrogate data retains the most important characteristics of the separate original variables (i.e., relative frequency and serial dependence), but breaks down the pairwise interdependence between the two variables, to be in line with the null hypothesis. The two generation mechanisms implemented in ConNEcT, a permutation-based and a model-based mechanism, are discussed in the following two subsections. Next, the same contingency measure is computed for each of the N by N surrogate variable pairs, yielding a sampling distribution of the contingency measure under the null hypothesis (step 4). In the fifth and last step, we compare this sampling distribution of the contingency values for the surrogate data with the value obtained in step 2. We consider the latter to be significantly stronger if it exceeds a certain percentile of the distribution, with the chosen percentile reflecting the adopted significance level.
Permutation-based data generation
As illustrated by Fig. 10, the permutation-based approach cuts the original variables into tenFootnote 5 roughly equal-sized segments of adjacent time points and subsequently permutes these segments (Moulder et al., 2018). By permutating the variables independently, the pairwise association between them is broken down. The relative frequency is left untouched and the serial dependence is also largely kept because sequences within the variables are often moved as a whole.
Schematic representation of the permutation-based data generation. The two observed time series (Variable 1 and Variable 2) are first cut into ten rather equally sized segments. These segments are randomly rearranged per variable yielding 100 permuted versions. To distinguish the solutions, we colored the second, fourth, sixth, and tenth segment in different shades of grey. The contingency measure of interest is computed for each pair of permuted variables.
Model-based data generation
In the model approach, the surrogate data is generated by simulating new data using characteristics of the original data. Specifically, the score for the first time point of the variable X is sampled from a Bernoulli distribution using the relative frequency \({p}_1^X\) as parameter. Next, all other scores Xt are drawn from a Bernoulli distribution using the observed conditional probability \({p}_{1\mid 0}^X\) if Xt-1=0 or the conditional probability \({p}_{1\mid 1}^X\) if Xt-1=1. The scores on Y are generated correspondingly. Note that the relative frequencies and auto-dependence of the thus generated data may differ somewhat from the intended values due to sampling fluctuations.
Differences between the two data generating mechanisms
Both generation mechanisms often, but not always lead to the same test result. These differences can be understood by realizing that the permutation-based test keeps the relative frequency exactly the same for all surrogate data, whereas the model-based test generated surrogate data show some variation in relative frequency and auto-dependence. On the one hand, the choice of which test to use should, therefore, be based on whether or not we consider the relative frequency of the observed data to be fixed or variable. On the other hand, the choice may also have practical implications. The model-based generation will yield a wider range of contingency values for the surrogate data, but also a smoother sampling distribution, due to the variation in relative frequency across the surrogate samples, as is illustrated in Fig. 11. Yet, in contrast to the permutation-based generation, this feature may generate surrogate data without observations for variables with a low relative frequency. However, if both variables have a relative frequency of 0, many contingency measures cannot be calculated (see Module 2). It is, therefore, good practice to take a critical look at the sampling distributions of the significance test.
Tutorial
Basics of the conTest function
Both significance tests are implemented in the conTest function. Like the conMx function, this function requires the specification of data, lag, conFun (see Module 2). Moreover, one should also set the following parameters:
-
typeOfTest: specifies whether a model-based (‘model’) or permutation-based (‘permut’; default) data generation mechanism is used
-
adCor: Logical parameter indicating whether (TRUE) or not (FALSE) the test should correct for auto-dependency, with TRUE being the default. If this parameter is set to FALSE, the test does not correct for serial dependence, because the surrogate data are generated based on the relative frequency only or by using segments of size 1. Although Bodner et al. (2021) showed that using these tests with time series data leads to an inflation of the type 1 error, we provide the option for comprehensiveness.
-
nBlox: the number of segments used in the permutation-based test; the default value of this parameter equals 10
-
nReps indicates the number of replicates or surrogate samples that are generated for each variable (default=100).
The output of the function is an S3 object of class ‘conTest’, containing:
-
allLinks: contingency values (see conMx output)
-
percentile: percentile of the sampling distribution where the observed contingency value is located
-
pValue: p-value for the one-sided significance test calculated as 1 minus the percentile.
-
para: the parameter settings for
-
typeOfTest
-
adCor
-
nBlox
-
nReps
-
funName
-
lag
-
varNames
-
-
samples: the generated surrogate samples per $variable1Name$variable2Name combination
Applying the significance test
We will illustrate the usage of the function employing the Symptom Data on the not-lagged data (default lag=0). We apply the permutation-based test (typeOfTest = ‘permut’), correcting for serial dependence (default adCor = TRUE) to the observed classic Jaccard values (conFun = funClassJacc). For each variable in the observed data set 100 replicates (nReps=100) of surrogate data will be generated. On all possible pairs of the surrogate data, the contingency measures are calculated, resulting in a sampling distribution consisting of 10,000 values. From the output test.result, we retrieve the classic Jaccard values in the observed data (test.result$allLinks) and the one-sided upper p-value (test.result$pValue).

From this output, we may, for example, conclude that the concurrent contingency between Guilt and Death has a Jaccard of value .63, which is significant with a p-value of .0003Footnote 6. The function returns the exact p-value, without correction for multiple testing, allowing the user to implement a multiple testing correction of choice.
Taking a closer look at the sampling distribution
As was already mentioned above, it is good practice to critically inspect the obtained sampling distribution. In our example, we might be interested in the concurrent relationships between Energy, Guilt, and Death, implying that we want to test three observed contingency values. To plot the associated matrix of sampling distributions, we first generate the corresponding conTest object. Second, we apply the hist.conTest functionFootnote 7. In the case of concurrent data, the distributions in the upper triangle of the matrix are exactly the same as those in the lower triangle for lagged data, they differ of course. By default, the 95% percentile is added to each histogram as a dashed line. Other percentiles can be requested by specifying the parameter signLev. The observed contingency value is added as a full grey line (Fig. 12).

Module 4: Networks
Theory
Up to now, we have examined data characteristics, quantified concurrent and temporal relationships between the variables over time and investigated whether these relationships are significant. This final module focuses on depicting these results as a network. We will point towards some advantages of collecting the bivariate relations in a network and give some ideas about how the network could be analyzed.
The added value of drawing a network
The variables under consideration constitute the nodes of the ConNEcT networks, with the size reflecting their relative frequencies (Module 1). The edges between the nodes visualize the strength of the obtained contingency values for the different variable pairs (Module 2). Studying concurrent contingencies results in a network with undirected links, while focusing on temporal sequences leads to a network with directed links. Networks may include all contingency values or only the significant ones (Module 3). Networks are thus a perfect way to integrate all ConNEcT analyses in one plot.
Networks make it easier to discern contingency patterns that are sometimes difficult to spot in contingency matrices or heatmaps. Figure 13 illustrates this by showing the temporal sequences using a heatmap as well as a network. The strength of the sequencing (lag=1) was quantified using a corrected Jaccard. Positive values are depicted with blue full arrows and negative values with brown dashed arrows. The positive loops between negative child behavior (Cneg) and negative behavior of the mother (Mneg), between positive child behavior (Cpos) and positive maternal behavior (Mpos), and between positive child behavior (Cpos) and working on the task together (Togth) are easily traced in the network, whereas this is harder in the heatmap.
Visibility of contingency relations comparing a a heatmap and b a network figure. Contingency values are calculated using corrected Jaccard with a lag of one. Node size is adapted to relative frequency. Saturation of the edges is proportional to the value. Blueish full line arrows indicate positive Jaccard values, brownish dashed arrows negative values. Significant links are indicated with * (p < .05).
Network psychometrics
Networks can be described in various ways. Multiple descriptive measures have been put forward, such as the size of a network (i.e., the number of nodes) or centrality measures (see Epskamp et al., 2012). The latter measures have gained a lot of attention, especially in psychopathological symptom networks. As discussed by Bringmann et al. (2019), the interpretation of these measures is, however, not as straightforward as it seems.
Some authors (e.g., Golino & Epskamp, 2017) have suggested screening the network for meaningful subgraphs. Subgraphs are subsets of the network, containing nodes and the links that connect them. Interesting are dense subgraphs, in which the nodes are all strongly interconnected, while the connections between the different subgraphs are then less strong. Indeed, the occurrence of clear subgraphs might reveal the underlying dimensionality of the data (Golino & Epskamp, 2017). The symptom network in Fig. 1, for instance, has a size of 8 and falls apart into four componentsFootnote 8, one comprising five nodes that co-occur, and the other three pertaining to single nodes. In the affective family network in Fig. 14, the happy behaviors of all family members seem to form a dense subgraph. For the other non-zero connections it is difficult to discern which belong to a dense subgraph and which are more isolated. In the tutorial part, we will use this example to explain, how these dense subgraphs can be identified with a clustering method. Specifically, we will use the cluster walktrap algorithm (Pons & Latapy, 2005) from the igraph package (Csardi & Nepusz, 2006).
Tutorial
All the analysis steps that were explained in the previous modules (lagging the data, calculating contingencies, and testing for significance) can be executed separately, but they can also be executed at once using the conNEcT function, which additionally yields a network.
The conNEcT function
The conNEcT function has the same input arguments as the ConTest function (Module 3), the conMx function (Module 2) and the conData function (Module 1): data, lag, conFun, typeOfTest, adCor, nBlox, nReps. In addition, one should specify whether the significance test should be executed (test=TRUE; default: FALSE), and if so, with which significance level (signLev; default=0.05). The function returns an S3 object of class ‘conNEcT’, comprising the following fields:
-
allLinks: The obtained contingency value matrix
-
signLinks: A contingency value matrix that only includes the significant contingencies; all other contingency values are set to zero (if no significance test is executed this matrix contains only NAs)
-
pValue: A matrix holding the p-values for the one-sided significance test (if no significance test is executed, all p-values are set to NA)
-
para (Parameters): Saving the settings of lags, test, typeOfTest, adCor, nBlox, nReps, funName, varNames
-
probs: A table containing the relative frequencies and conditional probabilities of all variables (see Module 1).
Example
We show the usage of the conNEcT function by applying it to the Family Data. We calculate the classic Jaccard (conFun=funClassJacc) on lagged data (lag=1; default: lag=0) and execute a permutation-based significance test (test=TRUE and typeOfTest= ‘permut’, default) that accounts for serial dependence (adCor = TRUE, default) using 10 Segments (nBlox=10, default) and 100 replications (nReps=100, default) with a significance level of 5% (signLev=0.05, default).

Plotting the network
A conNEcT object can be plotted as a network by calling the qgraph.conNEcTFootnote 9 function. The plot shows each variable as a node. The node size is adapted to the relative frequency. Default only the significant links are shown (signOnly=TRUE, default) Footnote 10; to display all links set signOnly to FALSE. Auto-loops are often uninformative and can be suppressed by adding diag=F. Since the qgraph function is taken from the R package qgraph (Epskamp et al., 2012), all features from qgraph can be used to optimize the plot (see examples for the networks in Figs. 1, 13 and 14 in supplementary material https://osf.io/p5ywg/)

To look for clusters in a network, we can, for example, use the cluster_walktrap function (Pons & Latapy, 2005) from the igraph package(Csardi & Nepusz, 2006). Our application of the function makes use of the qgraph and the igraph package which need to be installed first. Then we run the conNEcT function on the data. Here we chose to rely on all links (not only the significant ones) and to remove the diagonals. Clustering the contingency matrix reveals that there are three clusters, each consisting of the three adjacent nodes.

As we might consider this result slightly suboptimal in that we still have to look up the names of the variables, we might be tempted to add the variable names in an automated way, for example, with the following code:

Conclusions
We introduced the R package ConNEcT that implements the contingency measures based network approach for binary intensive longitudinal data. We showed in four modules (1) how the data can be explored, (2) how to compute pairwise contingencies among the variables, (3) how to investigate the significance of these contingency values, and (4) how to plot them in a network. Additionally, we gave first answers to some open questions: what are the similarities and differences of the included contingency measures, should the data be lagged, and if so, how are we able to get an idea of the appropriate lag, and how to detect patterns in the final network.
Notes
The function barplot.conData is an extension of the generic barplot function. When applying barplot() to a conData object (i.e., the output of the conData function), R will automatically call barplot.conData. It automatically provides the horizontal barplot, which matches nicely with the line plots (Fig. 1a, Fig. 2) as it has the same Y-axis. The code for a vertical barplot can be found in the supplementary material.
Warrens (2008a) concludes in his dissertation that the seven most important coefficients with the most attractive properties include the classic Jaccard index (≈ Tanimoto), Sokal and Michener (≈Rand index and proportion of agreement), and Cohen’s kappa.
Main et al. use diagonal recurrence profiles (DRP) from recurrence quantification analysis. This method was originally designed to find the optimal lag in reoccurring patterns within variables. Main et al. translate this idea to analyze the lags in dyadic data and determine whether the binary micro-coded interactions show concurrent synchrony (lags close to 0) or more a turn-taking pattern (lags differ from 0).
The plot function here refers to the plot.conProf functions which is an extension of the generic plot function. When applying plot() to a conProf object (i.e., the output of the conProf function), R will automatically call plot.conProf.
The number of ten segments leads to a good trade-off between leaving auto-correlation intact and creating enough variability to avoid that the segments of the two variables end up in the same spot by chance (Bodner et al., 2021; Bulteel et al., 2018a). Using fewer segments would retain even more of the serial dependencies, but cause less variation in how the segments could be arranged in the permutation strategy, and vice versa.
The p-value depends on the sampling distribution and may, therefore, slightly differ when the significance test is run several times, using different seeds.
The function hist.conTest is an extension of the generic hist function. When applying hist() to an object of the class conTest (i.e., the output of the conTest function), R will automatically call hist.conTest.
Components are subgraphs that are totally seperated from each other.
Unfortuantelly, qgraph is not (yet) a generic function, which means that we cannot simply call qgraph with a conNEcT object to call qgraph.conNEcT.
If the significance test had not been called when generating the conNEcT function (test=FALSE), but the plot opts for only significant links option (signOnly=TRUE), a warning message is displayed.
References
Albatineh, A. N., & Niewiadomska-Bugaj, M. (2011). Correcting Jaccard and other similarity indices for chance agreement in cluster analysis. Advances in Data Analysis and Classification, 5(3), 179–200. https://doi.org/10.1007/s11634-011-0090-y
Bakeman, R., & Quera, V. (2011). Sequential analysis and observational methods for the behavioral sciences. Cambridge University Press.
Bakeman, R., McArthur, D., & Quera, V. (1996). Detecting group differences in sequential association using sampled permutations: Log odds, kappa, and phi compared. Behavior Research Methods, Instruments, & Computers, 28(3), 446–457. https://doi.org/10.3758/BF03200524
Bodner, N., & Ceulemans, E. (2021). ConNEcT: Contingency Measure-Based Networks for Binary Time Series (R-package version 0.7.26) [Computer software]. https://CRAN.R-project.org/package=ConNEcT
Bodner, N., Kuppens, P., Allen, N. B., Sheeber, L. B., & Ceulemans, E. (2018). Affective family interactions and their associations with adolescent depression: A dynamic network approach. Development and Psychopathology, 30(4), 1459–1473. https://doi.org/10.1017/S0954579417001699
Bodner, N., Bosmans, G., Sannen, J., Verhees, M., & Ceulemans, E. (2019). Unraveling middle childhood attachment-related behavior sequences using a micro-coding approach. PLOS ONE, 14(10), e0224372. https://doi.org/10.1371/journal.pone.0224372
Bodner, N., Tuerlinckx, F., Bosmans, G., & Ceulemans, E. (2021). Accounting for auto-dependency in binary dyadic time series data: A comparison of model- and permutation-based approaches for testing pairwise associations. British Journal of Mathematical and Statistical Psychology, 74(S1), 86–109. https://doi.org/10.1111/bmsp.12222
Bodner, N., Bringmann, L., Tuerlinckx, F., De Jonge, P., & Ceulemans, E. (in press). ConNEcT: A novel network approach for investigating the co-occurrence of binary psychopathological symptoms over time. Psychometrika. https://doi.org/10.1007/s11336-021-09765-2
Borsboom, D. (2008). Psychometric perspectives on diagnostic systems. Journal of Clinical Psychology, 64(9), 1089–1108. https://doi.org/10.1002/jclp.20503
Borsboom, D. (2017). A network theory of mental disorders. World Psychiatry, 16(1), 5–13. https://doi.org/10.1002/wps.20375
Borsboom, D., & Cramer, A. O. J. (2013). Network Analysis: An Integrative Approach to the Structure of Psychopathology. Annual Review of Clinical Psychology, 9(1), 91–121. https://doi.org/10.1146/annurev-clinpsy-050212-185608
Boschloo, L., van Borkulo C. D., Rhemtulla, M., Keyes, K. M., Borsboom, D., & Schoevers, R. A. (2015). The Network Structure of Symptoms of the Diagnostic and Statistical Manual of Mental Disorders. PLOS ONE, 10(9), e0137621. https://doi.org/10.1371/journal.pone.0137621
Brandt, M. J., Sibley, C. G., & Osborne, D. (2019). What Is Central to Political Belief System Networks? Personality and Social Psychology Bulletin, 45(9), 1352–1364. https://doi.org/10.1177/0146167218824354
Bringmann, L. F., Vissers, N., Wichers, M., Geschwind, N., Kuppens, P., Peeters, F., Borsboom, D., & Tuerlinckx, F. (2013). A Network Approach to Psychopathology: New Insights into Clinical Longitudinal Data. PLoS ONE, 8(4), e60188. https://doi.org/10.1371/journal.pone.0060188
Bringmann, L. F., Pe, M. L., Vissers, N., Ceulemans, E., Borsboom, D., Vanpaemel, W., Tuerlinckx, F., & Kuppens, P. (2016). Assessing Temporal Emotion Dynamics Using Networks. Assessment, 1073191116645909. https://doi.org/10.1177/1073191116645909
Bringmann, L. F., Elmer, T., Epskamp, S., Krause, R. W., Schoch, D., Wichers, M., Wigman, J. T. W., & Snippe, E. (2019). What do centrality measures measure in psychological networks? Journal of Abnormal Psychology, 128(8), 892–903. https://doi.org/10.1037/abn0000446
Brusco, M., Steinley, D., Hoffman, M., Davis-Stober, C., & Wasserman, S. (2019). On Ising models and algorithms for the construction of symptom networks in psychopathological research. Psychological Methods, 24(6), 735–753. https://doi.org/10.1037/met0000207
Brusco, M., Cradit, J. D., & Steinley, D. (2021). A comparison of 71 binary similarity coefficients: The effect of base rates. PLOS ONE, 16(4), e0247751. https://doi.org/10.1371/journal.pone.0247751
Bulteel, K., Tuerlinckx, F., Brose, A., & Ceulemans, E. (2016a). Using Raw VAR Regression Coefficients to Build Networks can be Misleading. Multivariate Behavioral Research, 51(2–3), 330–344. https://doi.org/10.1080/00273171.2016.1150151
Bulteel, K., Tuerlinckx, F., Brose, A., & Ceulemans, E. (2016b). Clustering Vector Autoregressive Models: Capturing Qualitative Differences in Within-Person Dynamics. Frontiers in Psychology, 7. https://doi.org/10.3389/fpsyg.2016.01540
Bulteel, K., Mestdagh, M., Tuerlinckx, F., & Ceulemans, E. (2018a). VAR(1) based models do not always outpredict AR(1) models in typical psychological applications. Psychological Methods, 23(4), 740–756. https://doi.org/10.1037/met0000178
Bulteel, K., Tuerlinckx, F., Brose, A., & Ceulemans, E. (2018b). Improved Insight into and Prediction of Network Dynamics by Combining VAR and Dimension Reduction. Multivariate Behavioral Research, 53(6), 853–875. https://doi.org/10.1080/00273171.2018.1516540
Coan, J. A., & Gottman, J. M. (2007). The Specific Affect Coding System (SPAFF). In J. A. Coan & J. J. B. Allen, Handbook of emotion elicitation and assessment (pp. 267–285). Oxford University Press.
Cohen, J. (1960). A Coefficient of Agreement for Nominal Scales. Educational and Psychological Measurement, 20(1), 37–46. https://doi.org/10.1177/001316446002000104
Cramer, A. O. J., Waldorp, L. J., Maas, H. L. J. van der, & Borsboom, D. (2010). Comorbidity: A network perspective. Behavioral and Brain Sciences, 33(2–3), 137–150. https://doi.org/10.1017/S0140525X09991567
Cramer, A. O. J., van Borkulo C. D., Giltay, E. J., van der Maas, H. L. J., Kendler, K. S., Scheffer, M., & Borsboom, D. (2016). Major Depression as a Complex Dynamic System. PLOS ONE, 11(12), e0167490. https://doi.org/10.1371/journal.pone.0167490
Csardi, G., & Nepusz, T. (2006). The igraph software package for complex network research. InterJournal, Complex Systems, 1695.
Dalege, J., Borsboom, D., van Harreveld, F., van den Berg, H., Conner, M., & van der Maas, H. L. J. (2016). Toward a formalized account of attitudes: The Causal Attitude Network (CAN) model. Psychological Review, 123(1), 2–22. https://doi.org/10.1037/a0039802
Dujardin, A., Santens, T., Braet, C., De Raedt, R., Vos, P., Maes, B., & Bosmans, G. (2016). Middle Childhood Support-Seeking Behavior During Stress: Links With Self-Reported Attachment and Future Depressive Symptoms. Child Development, 87(1), 326–340. https://doi.org/10.1111/cdev.12491
Epskamp, S., & Fried, E. I. (2018). A tutorial on regularized partial correlation networks. Psychological Methods, 23(4), 617–634.
Epskamp, S., Cramer, A. O. J., Waldorp, L. J., Schmittmann, V. D., & Borsboom, D. (2012). qgraph: Network Visualizations of Relationships in Psychometric Data. Journal of Statistical Software, 48(4), 1–18.
Epskamp, S., Borsboom, D., & Fried, E. I. (2018a). Estimating psychological networks and their accuracy: A tutorial paper. Behavior Research Methods, 50(1), 195–212. https://doi.org/10.3758/s13428-017-0862-1
Epskamp, S., Waldorp, L. J., Mõttus, R., & Borsboom, D. (2018b). The Gaussian Graphical Model in Cross-Sectional and Time-Series Data. Multivariate Behavioral Research, 53(4), 453–480. https://doi.org/10.1080/00273171.2018.1454823
Fried, E. I., van Borkulo C. D., Cramer, A. O. J., Boschloo, L., Schoevers, R. A., & Borsboom, D. (2017). Mental disorders as networks of problems: A review of recent insights. Social Psychiatry and Psychiatric Epidemiology, 52(1), 1–10. https://doi.org/10.1007/s00127-016-1319-z
Golino, H. F., & Epskamp, S. (2017). Exploratory graph analysis: A new approach for estimating the number of dimensions in psychological research. PLOS ONE, 12(6), e0174035. https://doi.org/10.1371/journal.pone.0174035
Gwet, K. L. (2014). Handbook of inter-rater reliability: The definitive guide to measuring the extent of agreement among raters ; [a handbook for researchers, practitioners, teachers & students] (4. ed). Advanced Analytics, LLC.
Gwet, K. L. (2019). IrrCAC: Computing Chance-Correcte Agreement Coefficients (CAC) version 1.0. https://CRAN.R-project.org/package=irrCAC
Hamaker, E., Asparouhov, T., Brose, A., Schmiedek, F., & Muthén, B. (2018). At the Frontiers of Modeling Intensive Longitudinal Data: Dynamic Structural Equation Models for the Affective Measurements from the COGITO Study. Multivariate Behavioral Research, 53(6), 820–841. https://doi.org/10.1080/00273171.2018.1446819
Haslbeck, J. M. B., & Waldorp, L. J. (2020). mgm: Estimating Time-Varying Mixed Graphical Models in High-Dimensional Data. Ar**v:1510.06871 [Stat]. http://arxiv.org/abs/1510.06871. Accessed 18 Feb 2020.
Holloway, E. L., Wampold, B. E., & Nelson, M. L. (1990). Use of a paradoxical intervention with a couple: An interactional analysis. Journal of Family Psychology, 3(4), 385–402. https://doi.org/10.1037/h0080552
Hops, H., Biglan, A., Tolman, A., Arthur, J., & Longoria, N. (1995). Living in Family Environments (LIFE) coding system: (Reference manual for coders). Oregon Research Institute.
Hosenfeld, B., Bos, E. H., Wardenaar, K. J., Conradi, H. J., van der Maas, H. L. J., Visser, I., & de Jonge, P. (2015). Major depressive disorder as a nonlinear dynamic system: Bimodality in the frequency distribution of depressive symptoms over time. BMC Psychiatry, 15(1), 222. https://doi.org/10.1186/s12888-015-0596-5
Hubert, L., & Arabie, P. (1985). Comparing partitions. Journal of Classification, 2(1), 193–218. https://doi.org/10.1007/BF01908075
Isvoranu, A.-M., Borsboom, D., van Os, J., & Guloksuz, S. (2016). A Network Approach to Environmental Impact in Psychotic Disorder: Brief Theoretical Framework. Schizophrenia Bulletin, 42(4), 870–873. https://doi.org/10.1093/schbul/sbw049
Jaccard, P. (1901). Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bulletin de La Société Vaudoise Des Sciences Naturelles, 37, 547–579.
Jaccard, P. (1912). The Distribution of the Flora in the Alpine Zone. New Phytologist, 11(2), 37–50. https://doi.org/10.1111/j.1469-8137.1912.tb05611.x
Kuismin, M. O., & Sillanpää, M. J. (2017). Estimation of covariance and precision matrix, network structure, and a view toward systems biology: Estimation of covariance and precision matrix, network structure, and a view toward systems biology. Wiley Interdisciplinary Reviews: Computational Statistics, 9(6), e1415. https://doi.org/10.1002/wics.1415
Lafit, G., Tuerlinckx, F., Myin-Germeys, I., & Ceulemans, E. (2019). A Partial Correlation Screening Approach for Controlling the False Positive Rate in Sparse Gaussian Graphical Models. Scientific Reports, 9(1), 17759. https://doi.org/10.1038/s41598-019-53795-x
Lloyd, B. P., Yoder, P. J., Tapp, J., & Staubitz, J. L. (2016). The relative accuracy and interpretability of five sequential analysis methods: A simulation study. Behavior Research Methods, 48(4), 1482–1491. https://doi.org/10.3758/s13428-015-0661-5
Main, A., Paxton, A., & Dale, R. (2016). An Exploratory Analysis of Emotion Dynamics Between Mothers and Adolescents During Conflict Discussions. Emotion, 16(6), 913–928. https://doi.org/10.1037/emo0000180
Moulder, R. G., Boker, S. M., Ramseyer, F., & Tschacher, W. (2018). Determining synchrony between behavioral time series: An application of surrogate data generation for establishing falsifiable null-hypotheses. Psychological Methods, 23(4), 757–773. https://doi.org/10.1037/met0000172
Munoz, S. R., & Bangdiwala, S. I. (1997). Interpretation of Kappa and B statistics measures of agreement. Journal of Applied Statistics, 24(1), 105–112. https://doi.org/10.1080/02664769723918
Pons, P., & Latapy, M. (2005). Computing Communities in Large Networks Using Random Walks. In Pinar Yolum, T. Güngör, F. Gürgen, & C. Özturan (Eds.), Computer and Information Sciences—ISCIS 2005 (pp. 284–293). Springer. https://doi.org/10.1007/11569596_31
Rand, W. M. (1971). Objective Criteria for the Evaluation of Clustering Methods. Journal of the American Statistical Association, 66(336), 846–850. https://doi.org/10.1080/01621459.1971.10482356
Sheeber, L. B., Kuppens, P., Shortt, J. W., Katz, L. F., Davis, B., & Allen, N. B. (2012). Depression is associated with the escalation of adolescents’ dysphoric behavior during interactions with parents. Emotion, 12(5), 913–918. https://doi.org/10.1037/a0025784
Todeschini, R., Consonni, V., **ang, H., Holliday, J., Buscema, M., & Willett, P. (2012). Similarity Coefficients for Binary Chemoinformatics Data: Overview and Extended Comparison Using Simulated and Real Data Sets. Journal of Chemical Information and Modeling, 52(11), 2884–2901. https://doi.org/10.1021/ci300261r
van Borkulo, C. D., Borsboom, D., Epskamp, S., Blanken, T. F., Boschloo, L., Schoevers, R. A., & Waldorp, L. J. (2014). A new method for constructing networks from binary data. Scientific Reports, 4. https://doi.org/10.1038/srep05918
Van Keer, I., Ceulemans, E., Bodner, N., Vandesande, S., Van Leeuwen, K., & Maes, B. (2019). Parent-child interaction: A micro-level sequential approach in children with a significant cognitive and motor developmental delay. Research in Developmental Disabilities, 85, 172–186. https://doi.org/10.1016/j.ridd.2018.11.008
Warrens, M. (2008a). Similarity coefficients for binary data: Properties of coefficients, coefficient matrices, multi-way metrics and multivariate coefficients. Leiden University. https://scholarlypublications.universiteitleiden.nl/handle/1887/12987. Accessed 13 Sep 202.1
Warrens, M. (2008b). On Association Coefficients for 2×2 Tables and Properties That Do Not Depend on the Marginal Distributions. Psychometrika, 73(4), 777–789. https://doi.org/10.1007/s11336-008-9070-3
Wei, T., & Simko, V. (2021). R package ‘corrplot’: Visualization of a Correlation Matrix. (Version 0.88). https://github.com/taiyun/corrplot
Wilderjans, T. F., Lambrechts, G., Maes, B., & Ceulemans, E. (2014). Revealing interdyad differences in naturally occurring staff reactions to challenging behaviour of clients with severe or profound intellectual disabilities by means of Clusterwise Hierarchical Classes Analysis (HICLAS): Clusterwise HICLAS to detect interdyad differences. Journal of Intellectual Disability Research, 58(11), 1045–1059. https://doi.org/10.1111/jir.12076
Wongpakaran, N., Wongpakaran, T., Wedding, D., & Gwet, K. L. (2013). A comparison of Cohen’s Kappa and Gwet’s AC1 when calculating inter-rater reliability coefficients: A study conducted with personality disorder samples. BMC Medical Research Methodology, 13(1), 61. https://doi.org/10.1186/1471-2288-13-61
Yule, G. U. (1912). On the Methods of Measuring Association Between Two Attributes. Journal of the Royal Statistical Society, 75(6), 579. https://doi.org/10.2307/2340126
Acknowledgment
We thank Guy Bosmans (KU Leuven), Peter de Jonge (RU Groningen), and Lisa Sheeber (Oregon Research Institute) and their teams for collecting and sharing the data. The data is included in the R package ConNEcT. The research leading to the results reported in this paper was sponsored by research grants from the Research Council of KU Leuven (C14/19/054, PDM/20/062, iBOF/21/090). Supplementary material can be consulted at the OSF platform https://osf.io/p5ywg/.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Supplementary Information
ESM 1
(DOCX 466 kb)
Rights and permissions
About this article
Cite this article
Bodner, N., Ceulemans, E. ConNEcT: An R package to build contingency measure-based networks on binary time series. Behav Res 55, 301–326 (2023). https://doi.org/10.3758/s13428-021-01760-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3758/s13428-021-01760-w