Abstract
Background
Type IV lupus nephritis (LNIV) is a severe disease characterized by diffuse proliferative lesions, and its prognosis is worse with cellular crescent (LNIV-CC) involvement. Urinary exosomes have been shown to reflect the degree of kidney injury. This study was aimed to identify non-invasive diagnostic markers for LNIV-CC. We analysed the expression profile of microRNAs (miRNAs) isolated from urinary exosomes in patients with LNIV-CC and LNIV, and healthy individuals using high-throughput sequencing.
Results
A total of 66 differentially expressed miRNAs were identified, which were significantly enriched in 15 signalling pathways. Bioinformatic analysis revealed a co-expression network of miRNAs, predicted transcription factors and target mRNAs. Expression of three miRNAs including miR-3135b, miR-654-5p, and miR-146a-5p were further analysed and validated by reverse transcription-quantitative polymerase chain reaction. ROC analysis suggested these as candidate biomarkers for LNIV-CC.
Conclusions
LNIV-CC has a unique miRNA expression profile of urinary exosome and complex regulatory network. miR-3135b, miR-654-5p and miR-146a-5p in urinary exosomes could be used as novel non-invasive diagnostic markers for LNIV-CC.
Similar content being viewed by others
Background
Type IV lupus nephritis (LNIV) is a severe disease characterized by diffuse proliferative lesions. The treatment regimens of LNIV are determined based on pathological features in accordance to the guidelines of “the Kidney Disease: Improving Global Outcomes” (KDIGO) [1]. Recently, it was demonstrated that cellular crescent (CC) in renal biopsy is closely associated with rapid renal failure [2, 3]. Therefore, the 2017 Oxford Classification of IgAN added MEST-C scoring for cellular/fibrous crescents to assess the significant impact of crescents on kidney diseases [4]. However, invasive biopsy cannot be performed frequently in patients with LNIV. Therefore, screening for reliable biomarkers is urgently needed for non-invasive diagnosis and long-term monitoring of LNIV.
Exosomes are small vesicles (diameter of 30–120 nm) containing microRNAs (miRNAs), mRNAs, and proteins [5, 6]. They are found in almost all biological fluids including urine [5, 7]. Exosomes play an important role in the homeostasis and progression of kidney disease, known to interact with RNA-binding proteins or package them for transport [8, 9]. Although some urinary miRNAs are derived from circulation, the majority of miRNAs are derived from nephron exosomes [10, 11]. Since exosomes can be excreted in all cells of the nephrons, analysis of exosomes can accurately reflect renal dysfunction and structural damage [5, 7]. Therefore, miRNAs in urinary exosomes are potential biomarkers for diagnosis and prognosis [10, 11]. Recent studies indicated distinct miRNA expression profiles of urinary exosomes in patients with focal segmental glomerulosclerosis, and Type I and Type II diabetic nephropathy [12, 13]. Moreover, the miR-29c expression level in urinary exosomes was found to predict early fibrosis in lupus nephritis [14].
In the present study, we investigated the expression profiles of miRNAs isolated from urinary exosomes in patients with LNIV-CC and LNIV (without CC), we validated three selected differentially expressed miRNAs by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and performed bioinformatic analysis on the differentially expressed miRNAs and receiver operating characteristic (ROC) analysis for the validated miRNAs.
Methods
Subjects
A total of 44 patients were enrolled at the Department of Nephrology, the First Hospital Affiliated to Army Medical University from February 2017 to January 2018, including 15 active LNIV, 15 active LNIV-CC and 14 inactive LNIV. A total of 13 healthy volunteers without systemic diseases and normal renal function were also recruited from the health centre of the hospital from February 2017 to January 2018. miRNA sequencing was performed on 5 cases of active LNIV, 5 cases of active LNIV-CC, 4 cases of inactive LNIV, and 3 cases of healthy volunteers. The qRT-PCR validation was performed on additional patients (including 10 active LNIV, 10 active LNIV-CC and 10 inactive LNIV) and healthy volunteers (10 cases). All patients with LNIV were diagnosed according to the KDIGO Clinical Practice Guidelines [1], treated and tested by renal biopsy. Inactive nephritis was defined as a proteinuria level of less than 0.5 g day−1 with stable renal function without active urinary sediments. The study was approved by the Ethics Committee of the First Hospital Affiliated to Army Medical University (Southwest Hospital) in accordance with the Helsinki Declaration amended in 2008. All participants signed informed consents.
Urine processing, urinary exosome isolation, and exosomal RNA extraction
Fresh first-morning urine samples (100 ml) were collected in sterile containers and processed within 1 h after collection. Exosomes were isolated from the urine specimens using differential centrifugation. Briefly, the urine sample was centrifuged at 2500×g for 30 min to remove the cells and debris, and then centrifuged at 17,000×g for 30 min at 4 °C to remove large membrane vesicles. The resulting supernatant was centrifuged (Optima™ MAX-XP Ultracentrifuge, MLA-50 rotor, Beckman Coulter, USA) at 200,000×g for 70 min at 4 °C to pellet the exosomes. The resulting pellets were resuspended in phosphate-buffered saline (PBS) and centrifuged at 200,000×g for 70 min at 4 °C. Finally, the putative exosome pellets were resuspended in PBS and stored at − 80 °C.
RNA was extracted from the exosomes using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s recommended protocol. Denatured exosomes were spiked with the normalization control, the synthetic Caenorhabditis elegans miRNA cel-miR-39 (RiboBio, Guangzhou, China), lacking the endogenous human urinary miRNA. The RNA concentration was determined using the Qubit™ 3.0 Fluorometer (Life Technologies, USA).
Structural characteristics of exosome
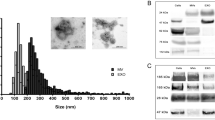
To verify the characteristics of exosomes, we examined the morphology of exosomes using transmission electron microscopy. A drop (~ 30 μl) of exosomes was resuspended in PBS and placed on parafilm. A formvar/carbon-coated copper grid was positioned on top of each drop for 15 min. Finally, a drop of 2% uranyl acetate was placed on the parafilm and incubated the exosomes for 5 min in the dark. The grids were negatively stained, air-dried, and examined with transmission electron microscopy (JEM-1400/JEM-1400 PLUS, Japan).
Western blot
The urinary exosomes were lysed in RIPA lysis buffer on ice, and then centrifuged at 12,000×g for 15 min. The supernatant was collected and transferred to a new EP tube. An equal amount of total soluble protein was subjected to 8% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (Life Technologies, USA) and transferred to a polyvinylidene fluoride membrane (Millipore, USA). The membranes were incubated with primary antibodies of CD9 and CD81 (CD9 rabbit, dilution 1:1000; CD81 rabbit, dilution 1:1000) and a primary antibody of non-exosomal protein calnexin mouse polyclonal (dilution 1:1000; Abcam, UK). Then, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies. Blots were visualized with chemiluminescence reagents (Beyotime, Shanghai, China).
miRNA sequencing
Total RNA was extracted from exosomes and used for miRNA high-throughput sequencing. Library preparation and miRNA sequencing were performed by Ribobio (Guangzhou, China). Briefly, total RNA samples were fractionated and only small RNAs were used for library preparation. After PCR amplification, the products were sequenced using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA) following the manufacturer’s instruction on running the instrument. Raw sequencing reads were obtained by using related Illumina’s analysis software.
Transcription factor (TF)–miRNA–mRNA network analysis
Differentially expressed exosomal miRNAs were screened out from the sequencing results. To predict TFs of the identified 66 differentially expressed miRNAs, BioPython [15] of JASPAR Database [16], FIMO [17] of MEME software [18], and Transcription Element Search System from the Computational Biology and Informatics Laboratory at the University of Pennsylvania were used. TargetScan (Release 7.2; http://www.targetscan.org/vert_72/), miRDB (http://mirdb.org/), miRTarBase (Release 7.0; http://mirtarbase.mbc.nctu.edu.tw/php/index.php), and miRWalk (Version 7.0; http://mirwalk.umm.uni-heidelberg.de/) software tools were used to predict the target gene of the identified differentially expressed miRNAs. Cytoscape (Version 3.6.1; http://www.cytoscape.org/) was used to establish miRNA-based regulatory network. The visualized network diagram of TFs, miRNAs, mRNAs, and their interactions was performed [19]. The enriched pathway analysis of the identified differentially expressed miRNAs was also performed with DIANA miRPath 3.0 [20].
RT-qPCR
The differentially expressed miRNAs were further validated by RT-qPCR. The expression of selected miRNAs was performed using the Stem-loop TaqMan miRNA RT Kit (Invitrogen, Shanghai, China) and TaqMan RT assay according to the manufacturer protocols. A fixed volume of cDNA (1.2 μl) was combined with TaqMan universal PCR master mix II no UNG. The stem-loop sequences were UGCCCAGGCUGGAGCGAGUGCAGUGGUGCAGUCAGUCCUAGCUCACUGCAGCCUCGAACUCCUGGGCU for miR-3135b, GGGUAAGUGGAAAGAUGGUGGGCCGCAGAACAUGUGCUGAGUUCGUGCCAUAUGUCUGCUGACCAUCACCUUUAGAAGCCC for miR-654a-5p, and CCGAUGUGUAUCCUCAGCUUUGAGAACUGAAUUCCAUGGGUUGUGUCAGUGUCAGACCUCUGAAAUUCAGUUCUUCAGCUGGGAUAUCUCUGUCAUCGU for miR-146a-5p. The relative expression levels of target miRNAs were compared between samples using the comparative cycle threshold (Ct) method 2(−ΔΔCt) after normalization to cel-miR-39 as an exogenous control.
Statistical analyses
Statistical analysis was performed by SPSS Statistics for Windows software version 19.0 (IBM, Armonk, NY, USA) with student’s t test. We measured the area under the curve (AUC) of the receiver operating characteristic (ROC) curve for the validated miRNAs. The discrimination capability of the simple risk score was also presented by ROC curve. p < 0.05 was considered statistically significant.
Results
Clinical characteristics of the study population
No significant difference was observed in the age, estimated glomerular filtration rate (eGFR) [21], and creatine levels among active LNIV, active LNIV-CC, inactive LNIV, and healthy controls. Active LNIV group had higher Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) scores and more 24-h proteinuria than inactive LNIV group (p < 0.01). Moreover, the 24-h proteinuria was lower in the active LNIV-CC group than the active LNIV group (p < 0.05) (Table 1).
Characteristics of isolated urinary exosomes
The extracted microvesicles were cup-shaped, < 100 nm in diameter, and scattered (Fig. 1a). Western blotting results showed positive expression of CD81 and CD9, and negative expression of calnexin (Fig. 1b), which is consistent with the typical characteristics of exosomes. Thus, the isolated microvesicles were exosomes.
Differential expression of exosomal miRNAs
A total of 904 altered exosomal miRNAs were detected between the LNIV-CC and control groups, including 53 with statistical significance (|log2 fold-change| > 1 and p < 0.05; Figs. 2 and 3). There were 662 common differentially expressed exosomal miRNAs detected between the LNIV-CC and LNIV (active and inactive) groups, including 66 with a significant difference (|log2 fold-change | > 1 and p < 0.05; Figs. 4 and 5). Among these differentially expressed miRNAs, a relative high expressed miR-3135b in LNIV (compared with other group), a relative high expressed miR-654-5p in LNIV-CC (compared with other group), and a not statistically significant differential expressed miR-146a-5p between LNIV-CC and LNIV (but higher than control group) were selected for validation with RT-qPCR.
Validation of differentially expressed exosomal miRNAs by RT-qPCR
For validation, RT-qPCR showed that the miR-3135b expression level in the active LNIV group was significantly higher than all other groups (p < 0.001), whereas the miR-654-5p expression level in the LNIV-CC group was significantly higher than LNIV and control groups (p < 0.001). The expression levels of miR-146a-5p in the inactive LNIV (p < 0.05) and LNIV-CC (p < 0.05) groups were significantly higher compared to that of the control group (Fig. 6). These results were consistent with the sequencing patterns, thereby confirming the accuracy and reliability of the sequencing data.
Pathway analysis of the differentially expressed exosomal miRNAs
DIANA miRPath (V3.0) [20] was used for pathway analysis of differentially expressed miRNAs between the LNIV-CC and LNIV groups with three databases including TargetScan, Tarbase 7.0, and microT-CDS v5.0. After obtaining the intersection of the results, 15 pathways related to crescents in LNIV were identified (Table 2), including Wingless–Int (Wnt) signalling pathway, signalling pathways regulating pluripotency of stem cells, focal adhesion, Hippo signalling pathway, neurotrophin signalling pathway, axon guidance, Forkhead-box Class O (FoxO) signalling pathway, ubiquitin-mediated proteolysis, epidermal growth factor receptor (ErbB or EGFR) signalling pathway, TGF-beta signalling pathway, arrhythmogenic right, ventricular cardiomyopathy, lysine degradation, N-Glycan biosynthesis, other types of O-glycan biosynthesis, and fatty acid biosynthesis. The number of identified miRNAs in those pathways were also shown (Table 2). Wnt signalling pathway, pathways regulating pluripotency of stem cells, focal adhesion, Hippo signalling pathway, neurotrophin signalling, axon guidance, and FoxO signalling pathway were correlated with more than 50 identified differentially expressed miRNAs, respectively.
TF–miRNA–mRNA network
The network diagram of TFs, miRNAs, mRNAs, and their interactions are shown in Fig. 7. Each miRNA node is connected to its TFs and targets with connecting lines. The more connections indicate a larger, more complicated and important miRNA node. Results indicated that the most important miRNAs included miR-1827, miR-16-5p, miR-671-5p, miR-324-5p, miR-552-3p, miR-22-5p, miR-513a-5p, miR-1231, miR-511-5p and miR-576-3p. This network diagram indicates important regulatory nodes in LNIV-CC.
Prediction value of miR-3135b, miR-654-5p, and miR-146a-5p
To evaluate the prediction value of miR-3135b, miR-654-5p, and miR-146a-5p, receiver operating characteristic (ROC) analysis was performed (Fig. 8). In LNIV-CC vs. LNIV, the specificity and sensitivity of miR-3135b were 93.33% and 83.33%, those of miR-654-5p were 63.33% and 96.67%, and those of miR-146-5p were 83.33% and 70%, respectively. In LNIV-CC vs. control, the specificity and sensitivity of miR-3135b were 40% and 93.33%, those of miR-654-5p were 60% and 90%, and those of miR-146-5p were 60% and 83.33%, respectively. Thus, miR-3135b, miR-654-5p, and miR-146a-5p were of significantly prediction values for LNIV-CC.
Discussion
miRNAs play an important role in biological function and renal pathology [12, 22]. Circulating miRNAs and urinary free miRNAs have been screened for diagnostic and therapeutic application in the renal pathology in lupus [23, 24]. However, those free miRNAs in body fluids are affected by enzymatic hydrolysis and therefore might not accurately reflect the pathological changes. Exosome is a new identified mode of intercellular communication. The miRNAs carried in exosomes can retain information on biological activity and quantity. Therefore, analysis of miRNAs in urinary exosomes might be more valuable. This study was the first to analyse the characteristics of miRNA expression in exosomes from LNIV-CC patients.
We demonstrated that the miRNA expression profile of urinary exosome in LNIV-CC is distinct from other groups, and miR-3135b and miR-654-5p are candidate biomarkers of LNIV-CC. The specificity of prediction of them was as high as 83.33–96.67%, which indicates the value of these miRNAs for further research and potential clinical application. miR-146a expression levels were also higher in the LNIV-CC group than the control group. miR-146a has been reported as a marker of glomerular and tubular interstitial tissue injury [25, 26]. The miR-146a expression level in lupus nephritis patients is not consistent. It was previously reported to be significantly higher in lupus nephritis patients than control group [27] but other studies showed that miR-146a is down-regulated in renal tissues and in mononuclear cells of patients with lupus nephritis compared to controls [28, 29]. Here, we did not detect a significant difference between the LNIV-CC and LNIV groups. It was suggested that miR-146a-5p likely does not play a major role in the regulation of CC that is characterized by substantial epithelial cell proliferation and monocyte infiltration.
miR-3135b is a biomarker of coronary artery calcification, heart failure, and non-ST segment elevation acute coronary syndromes, which is closely related to a poor cardiovascular prognosis [30, 31]. Thus, elevation of miR-3135b in the LNIV without crescents group may suggest that those patients are more likely to develop systemic lupus erythematosus-related cardiac damage. Our study firstly reports miR-3135b in association with kidney disease.
miR-654-5p has been reported to promote carcinoma proliferation in oral squamous cell through the miR-654-5p/GRAP/Ras/Erk signalling pathway [32], and down-regulation of miR-654-5p inhibited the proliferation of prostate cancer cells [33]. Thus, the high expression of miR-654-5p is closely related to the abnormal cell proliferation. CCs are formed because of the abnormal proliferation and more than three layers of epithelial cells. Since miR-654-5p was found to be highly expressed in the LNIV-CC group, it may be involved in the abnormal proliferation of epithelial cells.
There are several limitations in the present study. First, the number of subjects was relatively small. We only performed miRNA sequencing on only 17 patients, and validated the data on additional 40 patients. Therefore, large-scale cohort studies are needed to further analyse and validate these results, especially for diagnosis. Second, this is a cross-sectional study, and thus follow-up after treatment of lupus nephritis is needed to confirm the findings. Third, recent reports suggest that exosomes may cross the blood–brain barrier through receptor-mediated transcellular transport [34, 35]; however, it is currently not known whether this mechanism plays a role in the renal filtration barrier. Finally, the present study lacks functional experiments at cellular or molecular level to verify the causal relationship between miRNAs and lupus nephritis. We described the enriched signalling pathways with number of identified miRNAs in this study but the functional work regarding the miRNAs identified and the signalling pathways enriched should be performed in the future.
Overall, we demonstrate that LNIV with crescents has a unique miRNA expression profile of urinary exosome and complex regulatory network, and found that miR-3135b, miR-654-5p and miR-146a-5p in urinary exosomes could be used as novel non-invasive diagnostic markers for LNIV with crescents. Moreover, we identified 15 signalling pathways potentially associated with the formation of crescents in LNIV and established a visualized network of TF–miRNA–mRNA interactions, which lays the foundation for further studies on the pathology of lupus nephritis.
Abbreviations
- LNIV:
-
Type IV lupus nephritis
- LNIV-CC:
-
Type IV lupus nephritiscomplicated with cellular crescent
- miRNA:
-
microRNA
- TFs:
-
transcription factors
- RT-qPCR:
-
reverse transcription-quantitative polymerase chain reaction
- KDIGO:
-
“the Kidney Disease: Improving Global Outcomes”
- eGFR:
-
estimated glomerular filtration rate
- SLEDAI:
-
Systemic Lupus Erythematosus Disease Activity Index
- Cr:
-
creatine
- Wnt:
-
Wingless–Int
- FoxO:
-
Forkhead-box Class O
- ErbB or EGFR:
-
epidermal growth factor receptor
References
Radhakrishnan J, Cattran DC. The KDIGO practice guideline on glomerulonephritis: reading between the (guide)lines-application to the individual patient. Kidney Int. 2012;82:840–56.
Zhang X, Shi S, Ouyang Y, Yang M, Shi M, Pan X, et al. A validation study of crescents in predicting ESRD in patients with IgA nephropathy. J Transl Med. 2018;16:115.
Cai F, Han F, Wang H, Han H, Le J, Lan L, et al. The crescentic implication of renal outcomes in proliferative Lupus Nephritis. J Rheumatol. 2018;45:513–20.
Trimarchi H, Barratt J, Cattran DC, Cook HT, Coppo R, Haas M, et al. Oxford classification of IgA nephropathy 2016: an update from the IgA Nephropathy Classification Working Group. Kidney Int. 2017;91:1014–21.
Oeyen E, Van Mol K, Baggerman G, Willems H, Boonen K, Rolfo C, et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J Extracell Vesicles. 2018;7:1490143.
Yang Y, Hong Y, Cho E, Kim GB, Kim IS. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J Extracell Vesicles. 2018;7:1440131.
Krause M, Rak-Raszewska A, Naillat F, Searela U, Schmidt C, Ronkainen VP, et al. Exosomes as secondary inductive signals involved in kidney organogenesis. J Extracell Vesicles. 2018;7:1422675.
Lv LL, Cao YH, Pan MM, Liu H, Tang RN, Ma KL, et al. CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clin Chim Acta. 2014;428:26–31.
Keri KC, Regner KR, Dall AT, Park F. Urinary exosomal expression of activator of G protein signaling 3 in polycystic kidney disease. BMC Res Notes. 2018;11:359.
van Balkom BW, Pisitkun T, Verhaar MC, Knepper MA. Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int. 2011;80:1138–45.
Fang DY, King HW, Li JY, Gleadle JM. Exosomes and the kidney: blaming the messenger. Nephrology (Carlton). 2013;18:1–10.
Ramezani A, Devaney JM, Cohen S, Wing MR, Scott R, Knoblach S, et al. Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: a pilot study. Eur J Clin Invest. 2015;45:394–404.
Delić D, Eisele C, Schmid R, Baum P, Wiech F, Gerl M, et al. Urinary exosomal miRNA signature in Type II diabetic nephropathy patients. PLoS ONE. 2016;11:e0150154.
Solé C, Cortés-Hernández J, Felip ML, Vidal M, Ordi-Ros J. miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol Dial Transplant. 2015;30:1488–96.
Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–3.
Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4.
Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8.
Kutmon M, Kelder T, Mandaviya P, Evelo CTA, Coort SL. CyTargetLinker: a cytoscape app to integrate regulatory interactions in network analysis. PLoS ONE. 2013;8:e82160.
Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–6.
Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–70.
Zeng Y, Liu JX, Yan ZP, Yao XH, Liu XH. Potential microRNA biomarkers for acute ischemic stroke. Int J Mol Med. 2015;36:1639–47.
Navarro-Quiroz E, Pacheco-Lugo L, Lorenzi H, Díaz-Olmos Y, Almendrales L, Rico E, et al. High-throughput sequencing reveals circulating miRNAs as potential biomarkers of kidney damage in patients with systemic lupus erythematosus. PLoS ONE. 2016;11:e0166202.
Kaga H, Komatsuda A, Omokawa A, Ito M, Teshima K, Tagawa H, et al. Downregulated expression of miR-155, miR-17, and miR-181b, and upregulated expression of activation-induced cytidine deaminase and interferon-α in PBMCs from patients with SLE. Mod Rheumatol. 2015;25:865–70.
Ramachandran K, Saikumar J, Bijol V, Koyner JL, Qian J, Betensky RA, et al. Human miRNome profiling identifies MicroRNAs differentially present in the urine after kidney injury. Clin Chem. 2013;59:1742–52.
Lu J, Kwan BC, Lai FM, Tam LS, Li EK, Chow KM, et al. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology (Carlton). 2012;17:346–51.
Perez-Hernandez J, Forner MJ, Pinto C, Chaves FJ, Cortes R, Redon J. Increased urinary exosomal MicroRNAs in patients with systemic lupus erythematosus. PLoS ONE. 2015;10:e0138618.
Zheng CZ, Shu YB, Luo YL, Luo J. The role of miR-146a in modulating TRAF6-induced inflammation during lupus nephritis. Eur Rev Med Pharmacol Sci. 2017;21:1041–8.
Zhu Y, Xue Z, Di L. Regulation of MiR-146a and TRAF6 in the diagnose of lupus nephritis. Med Sci Monit. 2017;23:2550–7.
Liu W, Ling S, Sun W, Liu T, Li Y, Zhong G, et al. Circulating microRNAs correlated with the level of coronary artery calcification in symptomatic patients. Sci Rep. 2015;5:16099.
Wang H, Chen F, Tong J, Li Y, Cai J, Wang Y, et al. Circulating microRNAs as novel biomarkers for dilated cardiomyopathy. Cardiol J. 2016;24:65–73.
Lu M, Wang C, Chen W, Mao C, Wang J. miR-654-5p targets GRAP to promote proliferation, metastasis, and chemoresistance of oral squamous cell carcinoma through Ras/MAPK signaling. DNA Cell Biol. 2018;37:381–8.
Östling P, Leivonen SK, Aakula A, Kohonen P, Mäkelä R, Hagman Z, et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–67.
Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5.
El Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–57.
Authors’ contributions
LY contributed extensively to all aspects of the work presented in this manuscript. KZ and ZC participated in the conception and design of the project, and in the sample collection, data analysis and writing the manuscript. XX, XT, XB, BS, HZ and SL were involved in sample collection and data analysis. All authors read and approved the final manuscript.
Acknowledgements
We appreciate Jiabiao Chen who provided help in urine processing and support for the present study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The authors declare that data analyzed in the current study are available within the article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the First Hospital Affiliated to Army Medical University (Southwest Hospital) in accordance with the Helsinki Declaration amended in 2008. All participants were verbally informed of the purpose and content of the study and were enrolled after getting their signed informed consents.
Funding
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, Y., Xu, X., Tang, X. et al. MicroRNA expression profile of urinary exosomes in Type IV lupus nephritis complicated by cellular crescent. J of Biol Res-Thessaloniki 25, 16 (2018). https://doi.org/10.1186/s40709-018-0088-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40709-018-0088-0