Abstract
Lymphocyte-activation gene 3 (LAG3) is a highly anticipated immune checkpoint in the context of cancer, exerting regulatory control over immune cell proliferation and function to reinforce the advancement of cancers. However, the comprehensive functional analysis of LAG3 across various cancer types remains undisclosed; thus, this study aims to investigate the pan-cancer expression profile of LAG3. We have investigated the expression profile, prognostic significance, and genetic alterations of LAG3 in various cancers while elucidating its characteristic in immune response regulation. Our findings demonstrated that elevated LAG3 expression is significantly associated with favorable prognosis in patients with cutaneous melanoma (SKCM), and it may be a potential biomarker for SKCM. Furthermore, multiple immune algorithms have highlighted the important regulatory role of LAG3 for the tumor-infiltrating immune cells including CD8 + T cells, B cells, dendritic cells (DCs), macrophages, and natural killer (NK) cells. We also examined the distribution of LAG3 at the single-cell level and explored its functional significance. A comprehensive and systematic analysis of LAG3 would facilitate a comprehensive evaluation of LAG3 in cancer biology and provide valuable insights for cancer management.
Similar content being viewed by others
Introduction
The abnormal changes in genes have been proved to participate in the occurrence and development of cancer. Exploring the cancer-related genes and their signaling pathways by pan-cancer analysis could contribute to the diagnosis and clinical treatment of human cancers [1, 2].
Lymphocyte-activation gene 3 (LAG3) is located on chromosome 12 (12p13.32) and encodes a type 1 transmembrane protein involved in tumor immune regulation and clinical aggressiveness [3, 4]. It is widely expressed on various immune cells, predominantly found on the surface of diverse T lymphocytes including CD4 + and CD8 + subsets, as well as other immune cell populations [5]. As an inhibitory receptor, LAG3 exerts its influence on tumor progression by modulating T cell activation and effector function [6]. However, as an immunosuppressive receptor, there is a complex interplay between LAG3 and the infiltration and functionality of immune cells. In breast cancer, elevated levels of LAG3 expression have been positively associated with multiple immune cell infiltrates [7]. The immunohistochemical staining in gastric cancer reveals a positive correlation between an increased number of LAG3-positive cells and patient prognosis [8]. Furthermore, LAG3 can also stimulate mature dendritic cells (DCs) to produce cytokines such as IL-12 and TNF-a [34067904]. Notably, the emerging animal experiments and clinical trials have demonstrated that blockade of LAG3 enhances the anti-tumor efficacy, underscoring its potential in cancer therapy [9,10,11]. Given the intricate expression pattern of LAG3 across different cancers, comprehensive exploration of this receptor from a pan-cancer perspective is imperative.
In this study, we comprehensively assessed the characteristics of LAG3 in various cancer types using a range of bioinformatics platforms. We conducted comparative analyses of differential expression profiles and gene alterations of LAG3 between cancer and normal tissues, and evaluated their prognostic significance on the prognostic values and immune responses.
Materials and methods
Gene expression analysis
We conducted an analysis of LAG3 expression profiles between tumor tissues and normal tissues using Tumor Immune Estimation Resource 2.0 (TIMER2.0). We utilized the Gene Expression Profiling Interactive Analysis 2 (GEPIA2) database to investigate the relationship between LAG3 expression and the pathological stage of TCGA tumors.
Immumohistochemical staining
20 pairs of paraffin-embedded tissues from patients diagnosed with cutaneous malignant melanoma or non-specific skin inflammation were included. The LAG3 rabbit-derived polyclonal antibody from Proteintech Group (29,548-1-AP) was utilized with a dilution ratio of 1:4000 for this experiment.
Genetic alteration evaluation
The cBio Cancer Genomics Portal (cBioPortal) an interactive algorithm containing multidimensional cancer genomics datasets, was employed to investigate the molecular profiles and clinical attributes of pan-cancer samples in terms of genomics. In this study, the cBioPortal platform was utilized to evaluate the genetic alterations of LAG3 in TCGA cancers, including the types of genetic alteration and mutation sites.
Survival analysis
The prognostic value of LAG3 expression in multiple cancers was assessed using the GEPIA2 database. Additionally, the cBioPortal platform was utilized to determine the survival significance of LAG3 genetic alterations in cancer patients, encompassing overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS) and progression-free survival (PFS).
Immune infiltration analysis
The TIMER2.0 software was employed to investigate the correlation between LAG3 expression and various tumor-infiltrating immune cells populations, including B cells, CD8 + T cells, DCs, macrophages, neutrophils, natural killer cells (NK cells) and monocytes. Furthermore, the Gene Set Enrichment Analysis (GSEA) algorithm available at ** of tumor microenvironment. In uveal melanoma (UM), there was a negative correlation between LAG3 expression and invasion and metastasis (Fig. 6B). Figure 6C presents the distribution pattern of LAG3 at the single-cell level with in RB and UM samples. Next, we further investigated the LAG3 expression in various SKCM-derived cell lines using the TISCH database. The data revealed that LAG3 was present in diverse immune cell populations, particularly CD8 + T cells (Fig. 7A). The detailed distribution of LAG3 in various immune cells within several datasets is shown in Fig. 7B. These findings indicated that LAG3 might participate in modulating the CD8 + T cell response during tumorigenesis.
The expression levels of LAG3 at a single-cell sequence level. A, B The CancerSEA tool displayed the relationship between LAG3 expression and different functional status across pan-cancers. C The t-SNE diagrams portrayed the distributions of LAG3 expression at single-cell levels from RB and UM samples
The association between LAG3 expression and tumor-infiltrating immune cells
Based on several algorithms from TIMER2.0 database, we investigated the correlation between tumor-infiltrating immune cells and LAG3 expression across various cancer types. As shown in Fig. 8, we found a positive association between LAG3 expression and tumor infiltration of CD8 + central memory T cell in CESC, KIRC, SKCM, OV, UCEC, etc. Additionally, LAG3 expression exhibited a positive correlation with infiltration of B cell in LUAD, PAAD, OV, SKCM, TGCT and UCEC. Furthermore, LAG3 expression was positively correlated with the infiltration of activated myeloid DCs and plasmacytoid DCs in most cancers, such as BLCA, HNSC, KIRC, OV, and UCEC. Moreover, M1 macrophage infiltration demonstrated a positive correlation with LAG3 expression in several cancers including BLCA, HNSC, KIRC, READ, OV, SKCM and UCEC. Interestingly, ESCA showed a positive correlation between infiltration of NK cell and LAG3 expression while BRCA, GBM and SKCM displayed the negative correlation. Notably, no significant correlations were observed between LAG3 expression and other immune cell infiltrations (Additional file 1: Fig S1).
Discussion
LAG3, locating on chromosome 12 (12p13.32) and encoding a type 1 transmembrane protein, has been proved to be involved in immune regulation and clinical aggressiveness of cancer [18]. In breast cancer and lung adenocarcinoma, the high expression of LAG3 is closely related to the increase of T cell infiltration [7]. LAG3 could also directly and competitively bind with pHLA-II, to inhibiting T cell activation by affecting the combination of CD4 and pHLA-II [19]. Here, we performed a comprehensive analysis of LAG3 through multiple bioinformatics platforms to explore the functional roles of LAG3 in pan-cancer.
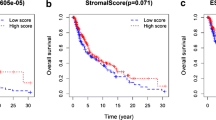
Based on the TCGA database, we investigated the expression of LAG3 in 33 cancers. We noticed that LAG3 was upregulated in most cancers, including BRCA, ESCA, GBM, HNSC, KIRC, LUAD, LURC, PCPG and STAD. Conversely, LAG3 was downregulated in COAD, KICH, LIHC, PRAD, READ, THCA and UCEC. Meanwhile, GEPIA2 database proved that in ACC, KIRC, COAD and OV, LAG3 expression was consistent with the pathological stage of patients. Then, Kaplan–Meier plotter showed that OV and UCEC patients with high LAG3 expression had better OS and RFS, while KIRC patients with high LAG3 expression had worse prognosis. These results suggest that the aberrantly expressed LAG3 could be a potential predictor for the prognosis of UCEC and OV cancer patients, and can assist in clinic pathological staging.
In addition, we also proved that the expression of LAG3 was significantly related with immune cell infiltration, including macrophage, CD8 + T cells, B cells, DCs, macrophage and NK cells. In earlier studies, LAG3 was a negative regulator of T cell activation and function [20]. Blocking the effect of LAG3 on human CD4 clones leads to increased production of IL-2, IL-4, IFN-γ, and TNF-α [21]. In mouse models of ovarian cancer, co-blocking LAG3 and PD-1 expressed on CD4 + and CD8 + TILs induces enhanced anti-tumor response [6]. Recent studies have shown that the combination of T cell checkpoint 41BB agonist and LAG3 antagonist could improve the anti-tumor immunity in pancreatic cancer [11]. Increased expression of LAG3 driven by IL6 led to decreased function of peripheral CD8 + T cells and reduced tumor therapeutic resistance [22]. Other studies also confirmed the regulatory roles of highly expressed LAG3 in the tumor infiltration of immune cells, including T cells, B cells, NK cells and DCs [23]. These results suggested the significant regulatory roles of LAG3 in the regulation of immune response.
Recent studies showed that high expression of LAG3 predicts a poor prognosis in patients due to its promotion of immune escape [24]. However, in SKCM, high expression of LAG3 suggests a favorable prognosis. These positive effects may be attributed to LAG3-mediated hyperactivation of immune system in SKCM [25, 26]. The discovery highlights the potential of LAG3 as a biomarker for assessing the immune system status in patients with SKCM, thereby offering valuable insights for its therapeutic management.
Conclusions
In conclusion, this is the first extensive research about the expression profiles, prognostic values, and functional significance of LAG3 in human pan-cancers. We demonstrate that LAG3 may be a novel prognostic marker for OV and UCEC. In addition, LAG3 expression could be associated with the tumor-infiltrating immune cells and might be an underlying predictive biomarker for cancer immunotherapeutic response. These results proved that LAG3 was closely related to tumor pathogenesis and immune response.
Availability of data and materials
The data used to support this article are included within the article.
References
Li Y, Lin Y, Aye L, Dong L, Zhang C, Chen F, Liu Y, Fan J, Gao Q, Lu H, et al. An integrative pan-cancer analysis of the molecular and biological features of glycosyltransferases. Clin Transl Med. 2022;12(7): e872.
Ren L, Yi J, Yang Y, Li W, Zheng X, Liu J, Li S, Yang H, Zhang Y, Ge B, et al. Systematic pan-cancer analysis identifies APOC1 as an immunological biomarker which regulates macrophage polarization and promotes tumor metastasis. Pharmacol Res. 2022;183: 106376.
Wang M, Du Q, ** J, Wei Y, Lu Y, Li Q. LAG3 and its emerging role in cancer immunotherapy. Clin Transl Med. 2021;11(3): e365.
Hu Y, Paris S, Bertolet G, Barsoumian HB, He K, Sezen D, Chen D, Wasley M, Silva JD, Mitchell JA, et al. Combining a nanoparticle-mediated immunoradiotherapy with dual blockade of LAG3 and TIGIT improves the treatment efficacy in anti-PD1 resistant lung cancer. J Nanobiotechnology. 2022;20(1):417.
Andrews LP, Marciscano AE, Drake CG, Vignali DA. LAG3 (CD223) as a cancer immunotherapy target. Immunol Rev. 2017;276(1):80–96.
Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, Eppolito C, Qian F, Lele S, Shrikant P, et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci U S A. 2010;107(17):7875–80.
Liu Q, Qi Y, Zhai J, Kong X, Wang X, Wang Z, Fang Y, Wang J. Molecular and clinical characterization of LAG3 in breast cancer through 2994 samples. Front Immunol. 2021;12: 599207.
Ulase D, Behrens HM, Kruger S, Heckl SM, Ebert U, Becker T, Rocken C. LAG3 in gastric cancer: it’s complicated. J Cancer Res Clin Oncol. 2023;149(12):10797–811.
Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutierrez E, Rutkowski P, Gogas HJ, Lao CD, De Menezes JJ, et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N Engl J Med. 2022;386(1):24–34.
Shao D, Chen Y, Huang H, Liu Y, Chen J, Zhu D, Zheng X, Chen L, Jiang J. LAG3 blockade coordinates with microwave ablation to promote CD8(+) T cell-mediated anti-tumor immunity. J Transl Med. 2022;20(1):433.
Gulhati P, Schalck A, Jiang S, Shang X, Wu CJ, Hou P, Ruiz SH, Soto LS, Parra E, Ying H, et al. Targeting T cell checkpoints 41BB and LAG3 and myeloid cell CXCR1/CXCR2 results in antitumor immunity and durable response in pancreatic cancer. Nat Cancer. 2023;4(1):62–80.
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–14.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4.
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605-d612.
Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47(D1):D900–8.
Han Y, Wang Y, Dong X, Sun D, Liu Z, Yue J, Wang H, Li T, Wang C. TISCH2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucleic Acids Res. 2023;51(D1):D1425–31.
Kraehenbuehl L, Weng CH, Eghbali S, Wolchok JD, Merghoub T. Enhancing immunotherapy in cancer by targeting emerging immunomodulatory pathways. Nat Rev Clin Oncol. 2022;19(1):37–50.
MacLachlan BJ, Mason GH, Greenshields-Watson A, Triebel F, Gallimore A, Cole DK, Godkin A. Molecular characterization of HLA class II binding to the LAG-3 T cell co-inhibitory receptor. Eur J Immunol. 2021;51(2):331–41.
Turnis ME, Andrews LP, Vignali DA. Inhibitory receptors as targets for cancer immunotherapy. Eur J Immunol. 2015;45(7):1892–905.
Huard B, Tournier M, Hercend T, Triebel F, Faure F. Lymphocyte-activation gene 3/major histocompatibility complex class II interaction modulates the antigenic response of CD4+ T lymphocytes. Eur J Immunol. 1994;24(12):3216–21.
Somasundaram A, Cillo AR, Lampenfeld C, Workman CJ, Kunning S, Oliveri L, Velez M, Joyce S, Calderon M, Dadey R, et al. Systemic immune dysfunction in cancer patients driven by IL6 induction of LAG3 in peripheral CD8+ T cells. Cancer Immunol Res. 2022;10(7):885–99.
Tu L, Guan R, Yang H, Zhou Y, Hong W, Ma L, Zhao G, Yu M. Assessment of the expression of the immune checkpoint molecules PD-1, CTLA4, TIM-3 and LAG-3 across different cancers in relation to treatment response, tumor-infiltrating immune cells and survival. Int J Cancer. 2020;147(2):423–39.
**a ZA, Lu C, Pan C, Li J, Li J, Mao Y, Sun L, He J. The expression profiles of signature genes from CD103(+)LAG3(+) tumour-infiltrating lymphocyte subsets predict breast cancer survival. BMC Med. 2023;21(1):268.
Workman CJ, Dugger KJ, Vignali DA. Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J Immunol. 2002;169(10):5392–5.
Chocarro L, Blanco E, Zuazo M, Arasanz H, Bocanegra A, Fernandez-Rubio L, Morente P, Fernandez-Hinojal G, Echaide M, Garnica M, et al. Understanding LAG-3 signaling. Int J Mol Sci. 2021;22(10):5282.
Acknowledgements
Not applicable.
Funding
This work was supported by the horizontal project (2022, 1 43010100; 2021–021, 143010100).
Author information
Authors and Affiliations
Contributions
PJ and ZZ designed this research. DZ performed the analysis. DZ wrote the original draft. SY prepared the figures. DZ and ZZ revised and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study reported in this manuscript was exempt from ethical committee approval because it was based on publicly available data.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
No significant correlation could be found between LAG3 expression and tumor infiltration of neutrophils and CD4 + T cells.
Additional file 2: Table S1.
The top 100 LAG3-related genes from the GEPIA2 database.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Peng, J., Du, Z., Sun, Y. et al. A combined analysis of multi-omics data reveals the prognostic values and immunotherapy response of LAG3 in human cancers. Eur J Med Res 28, 604 (2023). https://doi.org/10.1186/s40001-023-01583-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-023-01583-9