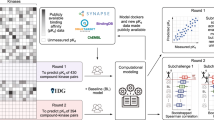
Abstract
Conventional machine learning (ML) and deep learning (DL) play a key role in the selectivity prediction of kinase inhibitors. A number of models based on available datasets can be used to predict the kinase profile of compounds, but there is still controversy about the advantages and disadvantages of ML and DL for such tasks. In this study, we constructed a comprehensive benchmark dataset of kinase inhibitors, involving in 141,086 unique compounds and 216,823 well-defined bioassay data points for 354 kinases. We then systematically compared the performance of 12 ML and DL methods on the kinase profiling prediction task. Extensive experimental results reveal that (1) Descriptor-based ML models generally slightly outperform fingerprint-based ML models in terms of predictive performance. RF as an ensemble learning approach displays the overall best predictive performance. (2) Single-task graph-based DL models are generally inferior to conventional descriptor- and fingerprint-based ML models, however, the corresponding multi-task models generally improves the average accuracy of kinase profile prediction. For example, the multi-task FP-GNN model outperforms the conventional descriptor- and fingerprint-based ML models with an average AUC of 0.807. (3) Fusion models based on voting and stacking methods can further improve the performance of the kinase profiling prediction task, specifically, RF::AtomPairs + FP2 + RDKitDes fusion model performs best with the highest average AUC value of 0.825 on the test sets. These findings provide useful information for guiding choices of the ML and DL methods for the kinase profiling prediction tasks. Finally, an online platform called KIPP (https://kipp.idruglab.cn) and python software are developed based on the best models to support the kinase profiling prediction, as well as various kinase inhibitor identification tasks including virtual screening, compound repositioning and target fishing.
Similar content being viewed by others
Introduction
The human kinome comprises more than 500 kinases, constituting approximately 1.7% of all human genes [1]. Protein kinases (PKs) play central roles in mediating most signaling pathways involved in cellular metabolism, transcription, cell cycle, apoptosis, and differentiation. Therefore, PKs have become one of the most interesting classes of drug targets for various diseases, including cancers [2,3,4], inflammation [5, 6], central nervous system disorders [7], cardiovascular diseases [8], complications of diabetes [9], and Alzheimer’s disease [10]. As such a significant class of targets, kinase inhibitors have been the focus of drug discovery. There are currently 71 FDA-approved small-molecule kinase inhibitors. In addition, approximately 110 innovative kinases are emerging as targets for drugs development in clinical trials [11]. Most FDA-approved drugs (63/71) targeting kinases are ATP-competitive inhibitors which inhibit kinases activity by binding to the ATP binding site of the kinase domain. However, the intrinsically highly conserved ATP binding sites of kinases may lead to off-target effects (i.e., low selectivity) of kinase inhibitors, potentially leading to undesirable side effects. Accordingly, identifying selective PK inhibitors remains an important challenge in the development of kinase-targeted drugs. Traditional kinase inhibitor assays are low-throughput methods that primarily measure the ability of compounds to reduce the phosphorylation activity for a given kinase (e.g. IC50) or their binding affinities to a kinase (dissociation constant, such as Ki and Kd). Notably, such measurement methods typically do not extend to the ability of a compound to inhibit the entire kinome. High-throughput kinase profiling assay has also become feasible in recent years, but the excessive cost makes it difficult to use as a routine early stage of drug discovery efforts [12].
Based on experimental data, a number of computational methods have been developed and published elsewhere, aiming to significantly reduce the cost, time and laborious involved in experimental identification. Generally, these computational methods can be classified into two major categories: structure- and ligand-based kinase inhibition and/or profiling prediction approaches (called virtual assay). Molecular docking, commonly used in structure-based prediction methods for kinase inhibition, has good generalizability, but its accuracy depends on the crystal structure of the kinase and the accuracy of the scoring function [13, 14]. Ligand-based methods include pharmacophore modelling, and quantitative structure–activity relationship (QSAR) [15,16,17,18,19,20,21]. Based on different kinase inhibitors-associated datasets, ML and DL algorithms such as naive Bayesian (NB) [22,60] were used to construct fingerprint-based predictive models. A set of 208 RDKit molecular descriptors (termed RDKitDes) was chosen for the development of descriptor-based predictive models. The fingerprints and descriptors were calculated using open source RDKit software (http://www.rdkit.org/, version: 2020.03.1).
In a molecular graph, the atomic and atomic pair features are used together as a feature matrix [61]. Chemprop and FP-GNN utilize RDKit software (version: 2020.09.5) to calculate molecular graphs. Other molecular graph-based representations were generated using DeepChem (version: 2.5.0). For example, the MolGraphConvFeatureizer module was used to calculate the molecular graphs for the GAT, MPNN, and Attentive FP models, while the ConvMolFeaturizer [62] module was used to compute the molecular graph representation for GCN models.
Selection of ML and DL algorithms for the assessment and model construction
Five mainstream ML and seven advanced DL algorithms were used to build the kinase profiling predictive modes for 354 kinases. These modelling methods (Table 1) are briefly introduced as follows.
Random forest (RF)
RF, developed by Svetnik et al.[42], is an ensemble recursive partitioning approach in which each recursive partitioning ‘tree’ is built from a bootstrapped sample of compounds, and each branch of a tree uses a random subset of descriptors [27]. The following five hyperparameters were tuned to achieve the optimal RF model: n_estimators (10–500), criterion (‘gini’ and ‘entropy’), max_depth (0–15), min_samples_leaf (1–10), and max_features (‘log2’, ‘auto’ and ‘sqrt’).
Naïve Bayesian (NB)
NB classifier is developed based on Bayes’ theorem [40] and widely used in molecular properties prediction and virtual screening (VS) projects [63,64,65,66]. Two hyperparameters were optimized for NB models construction: alpha (0.01–1) and binarize (0, 0.5, 0.8).
Support vector machine (SVM)
SVM was formally developed in 1995 [41] and quickly became a mainstream ML method due to its excellent performance in text classification tasks [67]. The principle of SVM is to determine the optimal hyperplane in the feature space by maximizing the boundaries between classes in N-dimensional space, which can distinguish objects with various class labels. Two hyperparmeters, Kernel coefficient (gamma, ‘auto’, 0.1–0.2) and penalty parameter C of the error term (C, from 1 to 100), were optimized for the development of SVM models.
K-nearest neighbor (KNN)
KNN is a commonly used supervised learning method with a simple mechanism. For a given test sample, it finds the k closest training samples in the training set based on distance measures (e.g., Manhattan, Euclidean, and Jaccard distance), and then makes a prediction based on the information of these k ‘neighbors’ [39]. In the training of KNN models, the default Euclidean distance metric was utilized, and three hyperparameters including n_neighbors (1–5), p (1–2), and weight function (‘uniform’, ‘distance’), were optimized.
Extreme gradient boosting (XGBoost)
XGBoost is one of the most representative ensemble ML algorithms under the gradient boosting framework [43]. It has been shown to achieve state-of-the-art (SOTA) performance on many standard classification benchmark datasets [37, 68, 69]. Seven hyperparameters were optimized: learning_rate (0.01–0.1), n_estimators (50–100), max_depth (3–5), min_child_weight (1–3), gamma (0–0.1), subsample (0.8–1.0), and colsample bytree (0.8–1.0).
Deep neural networks (DNN)
DNN is essentially an artificial neural network with an input layer, an output layer, and multiple hidden layers, which mimics the behavior of biological neural networks [44]. DNN consists of a large number of individual neurons [70, 71], and each neuron in the DNN architecture collects information from its associated neurons and a non-linear activation function was then used to activate the aggregated information. Three hyperparameters were optimized: dropouts (0.1, 0.2, 0.5), layer_sizes (64, 128, 256, 512) and weight_decay_penalty (0.01, 0.001, 0.0001).
Graph convolutional network (GCN)
GCN uses graph-structured data as features input [45], and consists of graph convolution layers, a readout layer, fully linked layers, and an output layer. The basic principle of GCN is to use edge information to aggregate node information, resulting in a new node representation. Several frameworks of GCN and variants have been proposed so far. For example, Duvenaud et al. [62] proposed a convolutional neural network that operates directly on molecular graphs, allowing end-to-end learning of prediction pipelines to exhibit better predictive performance for molecular property prediction tasks. Here, this GCN architecture was used to establish GCN models, and the following hyperparameters were optimized: weight decay (0, 10e-8, 10e-6, 10e-4), graph conv layers ([64, 64], [128, 128], [256, 256], learning rate (0.01, 0.001, 0.0001), and dense layer size (64, 128, 256).
Graph attention network (GAT)
GAT introduces an attention mechanism based on the GCN [46], which calculates the weights of the features of nodes and adjacent nodes through aggregation, and follows a self-aggregation strategy. GAT can better extract the spatial feature relationships of nodes compared to the GCN in the application of directed graphs [72]. Six hyperparameters were optimized in the training of the GAT models, including weight_decay (0, 10e-8, 10e-6, 10e-4), learning rate (0.01, 0.001, 0.0001), n_attention_heads (8, 16, 32), and dropouts (0, 0.1, 0.3, 0.5).
Message passing neural network (MPNN)
MPNN, first proposed by Gilmer and coworkers in 2017 [47], represents a commonly used GNN framework for various chemical prediction tasks. Many new GNN architectures have been developed based on the excellent performance and flexibility of MPNN framework for molecular property prediction [49, 73,74,75]. Herein, the main hyperparameters were optimized as follows: weight_decay (10e-8, 10e-6, 10e-4), learning rate (0.01, 0.001, 0.0001), graph_conv_layers ([64, 64], [128, 128], [256, 256]), num_layer_set2set (2, 3, 4), node_out_feats (16, 32, 64), and edge_hidden_feats (16, 32, 64).
Attentive FP
Attentive FP is an advanced GNN model that allows the model to focus on the most important elements of the input using graph attention mechanism [48]. It has been reported to exhibit SOTA performance for predicting molecular properties. Herein, the primary hyperparameters including dropout (0, 0.1, 0.5), graph feat size (50, 100, 200), num timesteps (1, 2, 3), num layers (2, 3, 4), learning rate (0.0001, 0.001, 0.01), and weight decay (0, 0.01, 0.0001), were optimized for the development of the Attentive FP models.
D-MPNN (Chemprop)
D-MPNN (Chemprop) was developed upon the MPNN framework by adopting a message-passing paradigm based on updating representations of directed bonds rather than atoms [49]. Chemprop has been successfully applied for the discovery of structurally distinct antibiotics [76]. Herein, the hyperparameters were optimized as follows: dropout (2, 3), dropout gat (0, 0.05), dim (1, 2), and gat scale (300, 400).
FP-GNN
Recently, FP-GNN as a novel DL architecture [50] was developed in our Lab for enhanced molecular properties prediction. FP-GNN not only learns to characterize the local atomic environment by propagating node information from nearby nodes to more distant nodes using the attention mechanism in a task-specific encoding, but also simultaneously learns a strong prior knowledge based on the fixed and complementary molecular fingerprints (MACCS, PubChem, and Pharmacophore ErG fingerprints). We used FP-GNN algorithm to build models for the kinase profiling prediction task. The hyperparameters were optimized as the following: dropout (0, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6), dropout gat (0, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6), dim (300, 350, 400, 450, 500, 550, 600), gat scale (0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8), nheads (2, 3, 4, 5, 6, 7, 8), and nhid (40, 45, 50, 55, 60, 65, 70, 75, 80).
The RF, SVM, KNN, and NB models were constructed using the Scikit-learn python package (https://github.com/scikit-learn/scikit-learn, version: 0.24.1) [77]; the XGBoost models were developed using the XGBoost python package (https://github.com/dmlc/xgboost, version: 1.3.3) [43]; four graph-based models (GCN, GAT, MPNN and Attentive FP) were established using the DeepChem python package (https://deepchem.io/); D-MPNN (Chemprop) models were constructed using the Chemprop python package (https://github.com/chemprop/chemprop); and FP-GNN models were developed using the FP-GNN software (https://github.com/idrugLab/FP-GNN). All ML and DL models were trained on CPU (Intel(R) Xeon(R) Silver 4216 CPU@2.10 GHz) and GPU (NVIDIA Corporation GV100GL [Tesla V100 PCIe 32 GB]), respectively. Additionally, Bayesian optimization was applied to optimize hyperparameters for FP-GNN and Chemprop models, while grid search method was employed to optimize hyperparameters for other models.
Performance evaluation metric
To benchmark the performance of different ML and DL tools for the kinase profiling prediction, six metrics, including specificity (SP/TNR), sensitivity (SE/TPR/Recall), Balanced accuracy (BA), F1 score, Matthew’s correlation coefficient (MCC), and area under the receiver operating characteristic (ROC) curve (AUC), are used and defined as follows:
where TP, TN, FP, and FN represent the number of true positives, true negatives, false positives, and false negatives, respectively.
AUC was the most commonly used criterion for kinase inhibitor activity prediction tasks [15, 29, 30, 34, 35, 78], we therefore selected AUC value as the indicator of the accuracy of the classification models for a fair comparison. Given that active compounds outnumbered inactive compounds in the current kinase profiling modelling dataset, with a positive-to-negative ratio of 3.83, F1 score was also utilized to judge the accuracy of the models [34, 79,80,81].
Results and discussion
Benchmark dataset analysis and model construction
We obtained a comprehensive kinase profiling modelling dataset from multiple sources by applying the criteria mentioned in the Methods section. This dataset contains 141,086 unique molecules involving in 216,823 inhibitory activity data points, which covers 354 kinases from nine groups in the human kinome: TK family (88 kinases), CMGC family (48 kinases), AGC family (44 kinases), CAMK family (46 kinases), STE family (38 kinases), TKL family (30 kinases), Atypical family (16 kinases), CK1 family (6 kinases), and Others (38 kinases), Detailed information of the dataset are shown in Additional file 2: Table S1. The average ratio of positive (actives) to negative (inactives) was approximately 3.83, implying that the modelling dataset is relatively unbalanced. Nonetheless, in order to objectively explore and evaluate the predictive performance of different computational methods, we preferred to utilize the raw data from experimentally validated molecules against these kinases, without adding theoretical decoys to deliberately balance the modelling dataset. Bemis–Murcko scaffold analysis was conducted to analyze the structural diversity of molecules in the dataset. The proportion of scaffolds to molecules for each kinase falls between 10 and 100%, with an average value of 51.0%, suggesting that the molecules of the dataset were structurally diverse. Besides, compounds have broad distributions of molecular weight (36.461–998.013) and AlogP (-8.895–11.509), indicating that the compounds in the modelling dataset have an extensive chemical space (Additional file 2: Table S2). Such results imply that the predictive models based on this dataset could exhibit better reliability and robustness.
For this comprehensive kinase profiling modelling dataset, a total of 148,680 classification predictive models were generated based on the three different types of molecular features using the selected 12 ML and DL algorithms. To fairly compare the performance of the ML and DL methods for the kinase profiling predictive task, the average of the evaluation metrics of the established models for each algorithm were calculated as the final result. The details of performance of the established models are described and discussed in the following sections.
Performance evaluation results of fingerprint-based ML and DL models
Five ML (KNN, NB, RF, SVM, and XGBoost) and one DL (DNN) approaches were used to build 106,200 predictive models based on five types of fingerprints (Morgan, MACCS, AtomPairs, FP2 and PharmacoPFP). Each model is denoted as a combination of the ML method and the corresponding molecular representation (e.g., DNN::Morgan).
As shown in Table 2, most of the fingerprint-based models performed well for the kinase profiling predictive task, with an average AUC value > 0.73 and average F1 value > 0.72 on the test sets. Despite the differences in the characteristics of the five molecular fingerprints, the RF method performed the best for 354 kinases (Fig. 2), with the highest average AUC value (0.769) and MCC value (0.395), and relatively high F1 score (0.731) and BA value (0.621). In addition, another ensemble learning methods, XGBoost, also showed considerable predictive performance, achieving the second highest AUC value (0.754) and F1 score (0.747), and relatively high BA value (0.651) and MCC value (0.367).
The Morgan fingerprints achieved highest mean AUC value (0.751 ± 0.035, Table 2), which implies that it is a relatively better molecular representation for kinase profiling prediction. In addition, combining different ML methods and different molecular fingerprints yielded different performance results, indicating that it is necessary to screen the combination of modelling algorithms and feature expressions to achieve the best performance. For example, the RF and XGBoost algorithm tends to use the FP2 fingerprints as input features to achieve the best model rather than the Morgan fingerprints. In contrast, the NB algorithm tends to utilize the Morgan fingerprints as input features to generate the best models rather than the FP2 fingerprints (Table 2).
We further analyzed the interval distribution of the average AUC values of the test sets of 354 kinase targets for each method. As shown in Fig. 3, although different combinations of fingerprints and modelling methods can produce different distributions of AUC values, statistical analysis found that the AUC values of the majority of the fingerprint-based models (~ 72.2%) were greater than 0.7. For example, the numbers of high quality (HQ, AUC > 0.7) for the RF::AtomPairs and XGBoost::AtomPairs models were 262 (Fig. 3A) and 248 (Fig. 3E) kinases, respectively. In addition, the RF::FP2 models showed obvious advantage, achieving the highest average AUC value (0.786 ± 0.150, Table 2). Importantly, it can achieve AUC values greater than 0.7 on 269 kinases (Fig. 3A).
The Morgan fingerprints owns the relatively better predictive performance with highest average AUC value, however, this does not necessarily mean that other fingerprints cannot outperform the Morgan fingerprints on individual kinases. Figure 4A showed that the FP2, AtomPairs, MACCS, and PharmacoPFP fingerprints contributed eight, eight, two, and two unique kinase targets in the models with an AUC ≥ 0.8. Although the Morgan fingerprints also contributed the most models with an AUC ≥ 0.8, and the majority of these models were commonly found by at least two of other four fingerprints (i.e. FP2, MACCS, Morgan and PharmacoPFP fingerprints). The most unique HQ models was obtained by the AtomPairs fingerprints with an average AUC greater than 0.9 (Fig. 4B), i.e. the FP2, MACCS, Morgan and PharmacoPFP fingerprints can generates two, three, six, and seven unique HQ models that cannot be obtained by the AtomPairs fingerprints.
Recently, Merget et al. [30] reported RF models based Morgan fingerprints for the profiling prediction of kinase inhibitors, with an average AUC of 0.76 on 291 kinases, and achieving HQ (AUC > 0.7) on ~ 200 kinases. Apparently, the RF::FP2 models proposed in this study are superior to the models from Merget et al. study in terms of the total of number of kinases (354) and the overall accuracy (mean AUC = 0.786), as well as the number of HQ models (269, AUC > 0.7). In addition, the RF::Morgan models proposed herein have comparable or superior performance to the models of Merget et al., i.e. it exhibited average AUC value of 0.774 on 354 kinases and achieved HQ models on 259 kinases. The results illustrated that the comprehensive kinase profiling dataset with large structural diversity and chemical space constructed in this paper is necessary for building robust and reliable kinase profiling prediction models, as well as the optimal combination of ML algorithms and molecular feature representations can help to develop more accurate models for the virtual profiling prediction of kinase inhibitors.
Performance evaluation results of descriptor-based ML and DL models
Subsequently, a total of 21,240 descriptor-based predictive models were successfully constructed and compared using the same modelling methods. The optimized RDKit-descriptors obtained using the SelectPercentile module (Percentile = 30) implemented in the scikit-learn package were utilized as input features for model construction. Detailed performance results of the descriptor-based models are listed in Additional file 2: Table S3. The average F1, AUC, and BA values for the test sets of these models are summarized in Table 3.
As shown Table 3, most descriptor-based predictive models performed quite well, with mean F1 scores = 0.74, and average AUC value greater than 0.75. In accordance with the fingerprint-based models evaluation results where RF method achieved the best performance, RF::RDKitDes also performed best with the highest average AUC value (0.798 ± 0.120) (Table 3) on these descriptor-based models, which by the way is higher than any other fingerprint-based models (Table 2). According to the average AUC values of these descriptor-based models (Table 3), KNN method achieved the second-ranked predictive performance, followed by NB and XGBoost methods.
Figure 5A illustrates that approximately 73% of the descriptor-based models are HQ models, which outperform the aforementioned fingerprint-based models. Taking the RF::RDKitDes model as an example, it not only achieved the highest mean AUC value, but achieved 288 HQ models (Fig. 5A) for 354 kinases. Clearly, the RF::RDKitDes model outperforms the corresponding RF-based fingerprint models in terms of both the average AUC metric and the number of HQ models (Table 2 and Fig. 3A), regardless of which molecular fingerprints is used as input features.
To further confirm whether descriptor-based models outperform fingerprint-based models, we systematically compare the evaluation metrics of these models. As shown in Fig. 5B, RDKitDes-based models slightly outperformed fingerprint-based models due to their best performances in terms of the high average F1 score, AUC, SE and MCC values. The detailed comparison results of descriptor- and fingerprint-based models for each ML algorithm are shown in Additional file 1: Fig. S1. For example, RDKitDes-based models achieved the highest F1 scores and AUC values on the RF, SVM, and KNN algorithms (Additional file 1: Figs. S1A, C and D), and slightly weaker and/or comparable performance on the NB, XGBoost and DNN methods (Additional file 1: Figs. S1B, E and F), when compared to fingerprint models based on these ML algorithms. These results highlighted that RDKitDes may be suitable for achieving the optimal performance of ML methods in the kinase profiling prediction task.
Performance evaluation results of graph-based DL models
Currently, various graph-based DL algorithms, which have recently been developed and achieved the SOTA performance in molecular property prediction tasks [ KIPP online platform is freely accessible at https://kipp.idruglab.cn/. Datasets and python version executable software of KIPP are freely available on Github: https://github.com/idrugLab/KinasePredictPro. Manning G, Whyte DB, Martinez R et al (2002) The protein kinase complement of the human genome. Science 298:1912–1934. https://doi.org/10.1126/science.1075762 Huang M, Shen A, Ding J, Geng M (2014) Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol Sci 35:41–50. https://doi.org/10.1016/j.tips.2013.11.004 Ma WW, Adjei AA (2009) Novel agents on the horizon for cancer therapy. CA Cancer J Clin 59:111–137. https://doi.org/10.3322/caac.20003 Sun C, Bernards R (2014) Feedback and redundancy in receptor tyrosine kinase signaling: relevance to cancer therapies. Trends Biochem Sci 39:465–474. https://doi.org/10.1016/j.tibs.2014.08.010 Clark JD, Flanagan ME, Telliez J-B (2014) Discovery and development of janus kinase (JAK) inhibitors for inflammatory diseases: miniperspective. J Med Chem 57:5023–5038. https://doi.org/10.1021/jm401490p Barnes PJ (2013) New anti-inflammatory targets for chronic obstructive pulmonary disease. Nat Rev Drug Discov 12:543–559. https://doi.org/10.1038/nrd4025 Muth F, Günther M, Bauer SM et al (2015) Tetra-substituted pyridinylimidazoles as dual inhibitors of p38α mitogen-activated protein kinase and c-Jun N-terminal kinase 3 for potential treatment of neurodegenerative diseases. J Med Chem 58:443–456. https://doi.org/10.1021/jm501557a Kikuchi R, Nakamura K, MacLauchlan S et al (2014) An antiangiogenic isoform of VEGF-A contributes to impaired vascularization in peripheral artery disease. Nat Med 20:1464–1471. https://doi.org/10.1038/nm.3703 Banks AS, McAllister FE, Camporez JPG et al (2015) An ERK/Cdk5 axis controls the diabetogenic actions of PPARγ. Nature 517:391–395. https://doi.org/10.1038/nature13887 Nygaard HB, van Dyck CH, Strittmatter SM (2014) Fyn kinase inhibition as a novel therapy for Alzheimer’s disease. Alzheimers Res Ther 6:8. https://doi.org/10.1186/alzrt238 Attwood MM, Fabbro D, Sokolov AV et al (2021) Author correction: trends in kinase drug discovery: targets, indications and inhibitor design. Nat Rev Drug Discov. https://doi.org/10.1038/s41573-021-00303-4 Goldstein DM, Gray NS, Zarrinkar PP (2008) High-throughput kinase profiling as a platform for drug discovery. Nat Rev Drug Discov 7:391–397. https://doi.org/10.1038/nrd2541 Li D-D, Meng X-F, Wang Q et al (2018) Consensus scoring model for the molecular docking study of mTOR kinase inhibitor. J Mol Graph Model 79:81–87. https://doi.org/10.1016/j.jmgm.2017.11.003 Burggraaff L, Lenselink EB, Jespers W et al (2020) Successive statistical and structure-based modeling to identify chemically novel kinase inhibitors. J Chem Inf Model 60:4283–4295. https://doi.org/10.1021/acs.jcim.9b01204 Kothiwale S, Borza C, Pozzi A, Meiler J (2017) Quantitative structure-activity relationship modeling of kinase selectivity profiles. Molecules 22:1576. https://doi.org/10.3390/molecules22091576 Kong Y, Yan A (2017) QSAR models for predicting the bioactivity of Polo-like kinase 1 inhibitors. Chemom Intell Lab Syst 167:214–225. https://doi.org/10.1016/j.chemolab.2017.06.011 Sciabola S, Stanton RV, Wittkopp S et al (2008) Predicting kinase selectivity profiles using free-Wilson QSAR analysis. J Chem Inf Model 48:1851–1867. https://doi.org/10.1021/ci800138n Sheridan RP, Nam K, Maiorov VN et al (2009) QSAR models for predicting the similarity in binding profiles for pairs of protein kinases and the variation of models between experimental data sets. J Chem Inf Model 49:1974–1985. https://doi.org/10.1021/ci900176y Hillisch A, Heinrich N, Wild H (2015) Computational chemistry in the pharmaceutical industry: from childhood to adolescence. ChemMedChem 10:1958–1962. https://doi.org/10.1002/cmdc.201500346 Keiser MJ, Roth BL, Armbruster BN et al (2007) Relating protein pharmacology by ligand chemistry. Nat Biotechnol 25:197–206. https://doi.org/10.1038/nbt1284 Keiser MJ, Setola V, Irwin JJ et al (2009) Predicting new molecular targets for known drugs. Nature 462:175–181. https://doi.org/10.1038/nature08506 Martin E, Mukherjee P, Sullivan D, Jansen J (2011) Profile-QSAR: a novel meta-qsar method that combines activities across the kinase family to accurately predict affinity, selectivity, and cellular activity. J Chem Inf Model 51:1942–1956. https://doi.org/10.1021/ci1005004 **a X, Maliski EG, Gallant P, Rogers D (2004) Classification of kinase inhibitors using a bayesian model. J Med Chem 47:4463–4470. https://doi.org/10.1021/jm0303195 Schürer SC, Muskal SM (2013) Kinome-wide activity modeling from diverse public high-quality data sets. J Chem Inf Model 53:27–38. https://doi.org/10.1021/ci300403k Lapins M, Wikberg JE (2010) Kinome-wide interaction modelling using alignment-based and alignment-independent approaches for kinase description and linear and non-linear data analysis techniques. BMC Bioinformatics 11:339. https://doi.org/10.1186/1471-2105-11-339 Niijima S, Shiraishi A, Okuno Y (2012) Dissecting kinase profiling data to predict activity and understand cross-reactivity of kinase inhibitors. J Chem Inf Model 52:901–912. https://doi.org/10.1021/ci200607f Chen B, Sheridan RP, Hornak V, Voigt JH (2012) Comparison of random forest and pipeline pilot naïve bayes in prospective QSAR predictions. J Chem Inf Model 52:792–803. https://doi.org/10.1021/ci200615h Cao D-S, Zhou G-H, Liu S et al (2013) Large-scale prediction of human kinase–inhibitor interactions using protein sequences and molecular topological structures. Anal Chim Acta 792:10–18. https://doi.org/10.1016/j.aca.2013.07.003 Bora A, Avram S, Ciucanu I et al (2016) Predictive models for fast and effective profiling of kinase inhibitors. J Chem Inf Model 56:895–905. https://doi.org/10.1021/acs.jcim.5b00646 Merget B, Turk S, Eid S et al (2017) Profiling prediction of kinase inhibitors: toward the virtual assay. J Med Chem 60:474–485. https://doi.org/10.1021/acs.jmedchem.6b01611 Yabuuchi H, Niijima S, Takematsu H et al (2011) Analysis of multiple compound–protein interactions reveals novel bioactive molecules. Mol Syst Biol 7:472. https://doi.org/10.1038/msb.2011.5 Unterthiner T, Mayr A, Klambauer G, et al. Deep Learning as an Opportunity in Virtual Screening. In: Workshop on Deep Learning and Representation Learning (NIPS2014). 2014. Li X, Li Z, Wu X et al (2020) Deep learning enhancing kinome-wide polypharmacology profiling: model construction and experiment validation. J Med Chem 63:8723–8737. https://doi.org/10.1021/acs.jmedchem.9b00855 Avram S, Bora A, Halip L, Curpăn R (2018) Modeling kinase inhibition using highly confident data sets. J Chem Inf Model 58:957–967. https://doi.org/10.1021/acs.jcim.7b00729 Li B, Lin M, Chen T, Wang L (2023) FG-BERT: a generalized and self-supervised functional group-based molecular representation learning framework for properties prediction. Brief Bioinform 24:bbad398. https://doi.org/10.1093/bib/bbad398 Wu Z, Jiang D, Hsieh C-Y et al (2021) Hyperbolic relational graph convolution networks plus: a simple but highly efficient QSAR-modeling method. Brief Bioinform 22:bbab112. https://doi.org/10.1093/bib/bbab112 Ye Q, Chai X, Jiang D et al (2021) Identification of active molecules against Mycobacterium tuberculosis through machine learning. Brief Bioinform 22:bbab068. https://doi.org/10.1093/bib/bbab068 Luukkonen S, Meijer E, Tricarico GA et al (2023) Large-scale modeling of sparse protein kinase activity data. J Chem Inf Model 63:3688–3696. https://doi.org/10.1021/acs.jcim.3c00132 Cover T, Hart P (1967) Nearest neighbor pattern classification. IEEE Trans Inf Theory 13:21–27. https://doi.org/10.1109/TIT.1967.1053964 Duda RO, Hart PE (1973) Pattern classification and scene analysis. Wiley, Hoboken. https://doi.org/10.1007/978-1-4471-0285-4 Zernov VV, Balakin KV, Ivaschenko AA et al (2003) Drug discovery using support vector machines. the case studies of drug-likeness, agrochemical-likeness, and enzyme inhibition predictions. J Chem Inf Comput Sci 43:2048–2056. https://doi.org/10.1021/ci0340916 Svetnik V, Liaw A, Tong C et al (2003) Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci 43:1947–1958. https://doi.org/10.1021/ci034160g Chen T, Guestrin C. Xgboost: A scalable tree boosting system//Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining. 2016: 785–794. https://doi.org/10.1145/2939672.2939785 McCulloch WS, Pitts W (1943) A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys 5:115–133. https://doi.org/10.1007/BF02478259 Kipf TN, Welling M. Semi-Supervised Classification with Graph Convolutional Networks. ar**v. 2017; 160902907 Veličković P, Cucurull G, Casanova A, et al. Graph Attention Networks. ar**v. 2018; 171010903 Gilmer J, Schoenholz SS, Riley PF, et al. Neural message passing for Quantum chemistry. In: Proceedings of the 34th International Conference on Machine Learning - Volume 70. JMLR.org, Sydney, NSW, Australia, pp 1263–1272. 2017. **ong Z, Wang D, Liu X et al (2020) Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J Med Chem 63:8749–8760. https://doi.org/10.1021/acs.jmedchem.9b00959 Yang K, Swanson K, ** W et al (2019) Analyzing learned molecular representations for property prediction. J Chem Inf Model 59:3370–3388. https://doi.org/10.1021/acs.jcim.9b00237 Cai H, Zhang H, Zhao D et al (2022) FP-GNN: a versatile deep learning architecture for enhanced molecular property prediction. Brief Bioinform 23(6):bbac408 Mendez D, Gaulton A, Bento AP et al (2019) ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 47:D930–D940. https://doi.org/10.1093/nar/gky1075 Kim S, Chen J, Cheng T et al (2021) PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 49:D1388–D1395. https://doi.org/10.1093/nar/gkaa971 Liu T, Lin Y, Wen X et al (2007) BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res 35:D198–D201. https://doi.org/10.1093/nar/gkl999 Sterling T, Irwin JJ (2015) ZINC 15—ligand discovery for everyone. J Chem Inf Model 55:2324–2337. https://doi.org/10.1021/acs.jcim.5b00559 Laufkötter O, Laufer S, Bajorath J (2020) Kinase inhibitor data set for systematic analysis of representative kinases across the human kinome. Data Brief 32:106189. https://doi.org/10.1016/j.dib.2020.106189 Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50:742–754. https://doi.org/10.1021/ci100050t Durant JL, Leland BA, Henry DR, Nourse JG (2002) Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci 42:1273–1280. https://doi.org/10.1021/ci010132r Carhart RE, Smith DH, Venkataraghavan R (1985) Atom pairs as molecular features in structure-activity studies: definition and applications. J Chem Inf Comput Sci 25:64–73. https://doi.org/10.1021/ci00046a002 O’Boyle NM, Banck M, James CA et al (2011) Open babel: an open chemical toolbox. J Cheminform 3:33. https://doi.org/10.1186/1758-2946-3-33 Gobbi A, Pop**er D (1998) Genetic optimization of combinatorial libraries. Biotechnol Bioeng 61:47–54. https://doi.org/10.1002/(SICI)1097-0290(199824)61:1%3c47::AID-BIT9%3e3.0.CO;2-Z Kearnes S, McCloskey K, Berndl M et al (2016) Molecular graph convolutions: moving beyond fingerprints. J Comput Aided Mol Des 30:595–608. https://doi.org/10.1007/s10822-016-9938-8 Duvenaud D, Maclaurin D, Aguilera-Iparraguirre J, et al. Convolutional Networks on Graphs for Learning Molecular Fingerprints. ar**v. 2015; 150909292 Wang L, Le X, Li L et al (2014) Discovering new agents active against methicillin-resistant staphylococcus aureus with ligand-based approaches. J Chem Inf Model 54:3186–3197. https://doi.org/10.1021/ci500253q Wang L, Chen L, Yu M et al (2016) Discovering new mTOR inhibitors for cancer treatment through virtual screening methods and in vitro assays. Sci Rep 6:18987. https://doi.org/10.1038/srep18987 Luo Y, Zeng R, Guo Q et al (2019) Identifying a novel anticancer agent with microtubule-stabilizing effects through computational cell-based bioactivity prediction models and bioassays. Org Biomol Chem 17:1519–1530. https://doi.org/10.1039/c8ob02193g Guo Q, Zhang H, Deng Y et al (2020) Ligand- and structural-based discovery of potential small molecules that target the colchicine site of tubulin for cancer treatment. Eur J Med Chem 196:112328. https://doi.org/10.1016/j.ejmech.2020.112328 Joachims T. Text categorization with support vector machines : learning with many relevant features. Proceedings of the ECML-98. 1998. Li S, Ding Y, Chen M et al (2021) HDAC3i-finder: a machine learning-based computational tool to screen for HDAC3 inhibitors. Mol Inform 40:2000105. https://doi.org/10.1002/minf.202000105 Jiang D, Wu Z, Hsieh C-Y et al (2021) Could graph neural networks learn better molecular representation for drug discovery? A comparison study of descriptor-based and graph-based models. J Cheminformatics 13:12. https://doi.org/10.1186/s13321-020-00479-8 Gawehn E, Hiss JA, Schneider G (2016) Deep learning in drug discovery. Mol Inform 35:3–14. https://doi.org/10.1002/minf.201501008 Ma J, Sheridan RP, Liaw A et al (2015) Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model 55:263–274. https://doi.org/10.1021/ci500747n Zhu L, Wan B, Li C et al (2021) Dyadic relational graph convolutional networks for skeleton-based human interaction recognition. Pattern Recognit 115:107920. https://doi.org/10.1016/j.patcog.2021.107920 Flam-Shepherd D, Wu T, Friederich P, Aspuru-Guzik A. Neural message passing on high order paths. ar**v. 2020; 200210413 Withnall M, Lindelöf E, Engkvist O, Chen H (2020) Building attention and edge message passing neural networks for bioactivity and physical–chemical property prediction. J Cheminform 12:1. https://doi.org/10.1186/s13321-019-0407-y Tang B, Kramer ST, Fang M et al (2020) A self-attention based message passing neural network for predicting molecular lipophilicity and aqueous solubility. J Cheminform 12:15. https://doi.org/10.1186/s13321-020-0414-z Stokes JM, Yang K, Swanson K et al (2020) A deep learning approach to antibiotic discovery. Cell 180:688-702.e13. https://doi.org/10.1016/j.cell.2020.01.021 Swami A, Jain R (2013) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830 Sorgenfrei FA, Fulle S, Merget B (2018) Kinome-wide profiling prediction of small molecules. ChemMedChem 13:495–499. https://doi.org/10.1002/cmdc.201700180 Abdelbaky I, Tayara H, Chong KT (2021) Prediction of kinase inhibitors binding modes with machine learning and reduced descriptor sets. Sci Rep 11:706. https://doi.org/10.1038/s41598-020-80758-4 Sánchez-Cruz N, Medina-Franco JL (2021) Epigenetic target fishing with accurate machine learning models. J Med Chem 64:8208–8220. https://doi.org/10.1021/acs.jmedchem.1c00020 Kc GB, Bocci G, Verma S et al (2021) A machine learning platform to estimate anti-SARS-CoV-2 activities. Nat Mach Intell 3:527–535. https://doi.org/10.1038/s42256-021-00335-w Wu Z, Ramsundar B, Feinberg EN et al (2018) MoleculeNet: a benchmark for molecular machine learning. Chem Sci 9:513–530. https://doi.org/10.1039/C7SC02664A Eid S, Turk S, Volkamer A et al (2017) KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics 18:1–6 Karaman MW, Herrgard S, Treiber DK et al (2008) A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 26:127–132. https://doi.org/10.1038/nbt1358 Graczyk PP (2007) Gini coefficient: a new way to express selectivity of kinase inhibitors against a family of kinases. J Med Chem 50:5773–5779. https://doi.org/10.1021/jm070562u Bland JM (2000) Statistics notes: the odds ratio. BMJ 320:1468–1468. https://doi.org/10.1136/bmj.320.7247.1468 Liang X, Lv F, Wang B et al (2017) Discovery of 2-((3-Acrylamido-4-methylphenyl)amino)-N-(2-methyl-5-(3,4,5-trimethoxybenzamido)phenyl)-4-(methylamino)pyrimidine-5-carboxamide (CHMFL-BMX-078) as a highly potent and selective type II irreversible bone marrow kinase in the X chromosome (BMX) kinase inhibitor. J Med Chem 60:1793–1816. https://doi.org/10.1021/acs.jmedchem.6b01413 We acknowledge the allocation time from the SCUTGrid at South China University of Technology. This work was supported in part by the National Natural Science Foundation of China (81973241) and the Natural Science Foundation of Guangdong Province (2020A1515010548). LW conceived and designed the project. JW and YC contributed to the literature search, data collection, and algorithm architecture realization. JW and DZ was response for analyzing the modelling results and implementation models to web-server. JH and ML were in charge of web-based software construction on front-end and back-end respectively. LW provided support and critically revised the manuscript. All authors have read and agreed to the published version of the manuscript. The authors declare no competing interests. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Detailed comparison performance of descriptor- and fingerprint-based models using various ML algorithms. (A), (B), (C), (D), (E), and (F) represent the comparison results for the RF, NB, SVM, KNN, XGB, and DNN methods, respectively. Fig S2. Comparison of average F1 scores of (A) AtomPairs-, (B) FP2-, (C) MACCS-, (D) Morgen-, (E) PharmacoPFP-, (F) RDKitDes-, and (G) Graph-based models. The assay-F1 scores for various ML algorithms are displayed as boxplot. Middle spheres represent the median, and boxes represents the interquartile range (IQR) from the median. Fig S3. Comparison of average BA values of (A) AtomPairs-, (B) FP2-, (C) MACCS-, (D) Morgen-, (E) PharmacoPFP-, (F) RDKitDes-, and (G) Graph-based models. The assay-BA values for various ML algorithms are displayed as boxplot. Middle spheres represent the median, and boxes represent the interquartile range (IQR) from the median. Fig S4. Comparison of average MCC values of (A) AtomPairs-, (B) FP2-, (C) MACCS-, (D) Morgen-, (E) PharmacoPFP-, and (F) RDKitDes-, (G) Graph-based models. The assay-MCC values for various ML algorithms are displayed as boxplot. Middle spheres represent the median, and boxes represent the interquartile range (IQR) from the median. Details on benchmark dataset for kinase profiling prediction task used in this study. Table S2. Structural diversity and chemical space analysis of the compounds in each kinase. Table S3. Detailed performance results of different ML methods. Table S4. Detailed individual kinases where the GCN models outperform the RF::RDKitDes models. Table S5. Detailed individual kinases where the FP-GNN models outperform the RF::RDKitDes models. Table S6. The optimal in silico predictive models for each kinase in terms of AUC metric. Table S7. Comparison performance of models based on combined features and single feature in terms of F1 score. Table S8. Ranking of all single models by AUC values. Table S9. Comparison of our models with the reported in silico prediction models for kinase profiling prediction task. Table S10. The predicted activity probability and experimental % activity of CHMFL-BMX-078. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data. Wu, J., Chen, Y., Wu, J. et al. Large-scale comparison of machine learning methods for profiling prediction of kinase inhibitors.
J Cheminform 16, 13 (2024). https://doi.org/10.1186/s13321-023-00799-5 Received: Accepted: Published: DOI: https://doi.org/10.1186/s13321-023-00799-5Data availability
References
Acknowledgements
Funding
Author information
Authors and Affiliations
Contributions
Corresponding author
Ethics declarations
Competing interests
Additional information
Publisher's Note
Supplementary Information
Additional file 1: Fig S1.
Additional file 2: Table S1.
Rights and permissions
About this article
Cite this article
Keywords