Abstract
Background
Continuous crop** challenges constrain the development of agriculture. Three main obstacles limit continuous crop**: autotoxicity of plant allelochemicals, deterioration of physicochemical characteristics of soil, and microflora imbalance. Plant-derived phenolic acids can cause autotoxicity, which is considered the main factor mediating continuous crop** obstacles. Reducing the phenolic acids in continuous crop** soils can decrease the autotoxicity of phenolic acids and ameliorate continuous crop** obstacles. Therefore, it is important to study the microbial resources that degrade allelochemical phenolic acids. Thus, the bacterial strain V4 that can degrade phenolic acids was isolated, identified, and genomically analyzed.
Results
Strain V4 isolated from strawberry soil using vanillic acid-mineral agar was identified as a Gram-negative short rod bacterium. Subsequent 16S rRNA phylogenetic analysis revealed that V4 clustered with members of the genus Sphingobium. The most closely related species were Sphingobium lactosutens DS20T (99% similarity) and Sphingobium abikonense NBRC 16140T (97.5% similarity). V4 also shared > 95% sequence similarity with other members of Sphingobium, so Sphingobium sp. V4 was named accordingly. Biochemical tests revealed that the biochemical characteristics of Sphingobium sp. V4 were similar to its most similar strains except for some properties. Sphingobium sp. V4 effectively degraded vanillic acid, ferulic acid, p-coumaric acid, p-hydroxybenzoic acid, and syringic acid. V4 grew best at the conditions of 30 °C, pH 6.0–7.0, and 0–0.05% NaCl. 500 mg/L vanillic acid was completely degraded by V4 within 24 h under the optimal conditions. Whole genome analysis showed that Sphingobium sp. V4 contained one chromosome and three plasmids. Two genes involved in vanillic acid degradation were found in the V4 genome: the gene encoding vanillate O-demethylase oxidoreductase VanB on the chromosome and the gene encoding vanillate monooxygenase on a large plasmid. The organization of vanillate catabolic genes differed from the adjacent organization of the genes, encoding vanillate o-demethylase VanA and VanB subunits, in Pseudomonas and Acinetobacter.
Conclusions
The isolated bacterium Sphingobium sp. V4 degraded multiple phenolic acids. Its properties and genome were further analyzed. The study provides support for further investigation and application of this phenolic acid-degrading microorganism to alleviate continuous crop** obstacles in agriculture.
Similar content being viewed by others
Background
Continuous crop** obstacles present a difficult problem in agricultural production worldwide. Many crops are prone to such continuous crop** obstacles, including peanut, soybean, tomato, cucumber, potato, strawberry, tobacco, and Panax notoginseng (Yu et al. 2000; Zhou et al. 2012; Li et al. 2016; Tan et al. 2017). The causes of continuous crop** obstacles are complex and generally related to the autotoxicity of plant allelochemicals, the deterioration of soil physical and chemical properties, and imbalances in soil microflora. Phenolic acids in plant root exudates and stubble decaying substances can cause allelopathy, which is the main mediating factor of continuous crop** difficulties (Chen et al. 2014; Wu et al. 2015; Bai et al. 2019). Levels of phenolic acids, such as p-coumaric acid, vanillic acid, p-hydroxybenzoic acid, and ferulic acid, are significantly higher in continuous crop** soils and have exhibited allelopathy which inhibits the growth of the same or related crops and increases the incidence of soil-borne diseases (Carlsen et al. 2009; Chen et al. 2014; Gulzar and Siddiqui 2015; Bai et al. 2019). Bai et al. reported that coumaric acid, p-hydroxybenzoic acid, vanillic acid, vanillin, and ferulic acid significantly accumulated with increased duration of continuous crop** for tobacco, and the accumulated phenolic acids caused strong allelopathic activity (Bai et al. 2019). Phenolic acids also affect the microbial community structure by acting as carbon and energy sources or as signaling molecules (She et al. 2017; Liu et al. 2017; ** soils helps crops by reducing the autotoxicity of phenolic acids and the occurrence of soil-borne diseases and, thus, alleviates continuous crop** obstacles. The published reports of bacteria that degrade allelochemical phenolic acids are increasing in number, not only for Pseudomonas, but also for Paraburkholderia and other genera (Mohan and Phale 2017; Wilhelm et al. 2020; Araki et al. 2020; Liu et al. 2023).
In this study, a Sphingobium sp. V4 was isolated, identified, and observed to degrade phenolic acids with high efficiency. Its confirmed substrates included vanillic acid, ferulic acid, p-coumaric acid, p-hydroxybenzoic acid, and syringic acid. Its whole genome and vanillic acid demethylase gene were also analyzed. This work provides a potential bacterial strain for the development of bacterial agents to prevent and control continuous crop** obstacles.
Methods
Strain isolation
Soil samples were collected from a strawberry field located in Zibo, Shandong Province, China, in 2018. The geographic coordinates of the strawberry field were 36° 53′ N and 118° 04′ E. Vanillic acid-mineral (VM) agar medium was used to isolate the vanillic acid-degrading bacterium. The composition of the VM liquid medium was as follows: 0.5 g of vanillic acid, 0.2 g of KNO3, 5 g of Na2HPO4 ∙12H2O, 3 g of KH2PO4, 0.5 g of NaCl, 0.003 g of CaCl2, 0.003 g of MgSO4∙7H2O, and 1000 mL of distilled water. VM agar medium was generated by adding 20 g/L of agar to the VM liquid medium. VM was supplemented with 0.5 g/L (w/v) of yeast extract to produce a VM yeast extract (VMY) medium. Sterilization was performed at 121 °C for 20 min in an autoclave. Soil sample suspensions of 10−2, 10−3, and 10−4 concentrations were made with sterilized distilled water. Then, 100 μL suspensions of each concentration were spread on the VM agar medium to isolate bacteria. Each concentration was replicated three times. The plates were incubated at 30 °C for 3 to 5 days, and bacterial colonies were observed daily. After the colonies grew well, the strains were isolated and purified on VM agar medium.
The purified colony was inoculated into VM liquid medium, and the grown cell culture was added to fresh VM liquid medium with a 10% (v/v) inoculation ratio to test its ability to degrade vanillic acid. Cultivation conditions were maintained at 30°C and 150 r/min for 5 days. Samples were collected at 0 and 5 days. The content of vanillic acid was determined by high-performance liquid chromatography (HPLC) (Shimadzu, Japan) with an XDB-C18 reversed-phase column (4.6 mm × 250 mm). For HPLC, 0.16% acetic acid and methanol (40:60) were used as the mobile phase at a flow rate of 1.0 mL/min. The column temperature was 30 °C, and the detector wavelength was 280 nm. The substrate degradation rate was calculated as [(initial substrate content − final substrate content)/initial substrate content] × 100%. A control was established in which 10% (v/v) sterilized distilled water was added instead of V4 culture. The degradation test was repeated three times.
Phylogeny of 16S rRNA gene sequences
The genomic DNA of the vanillic acid-degrading bacterium V4 was extracted using the Bacterial Genomic DNA Rapid Extraction Kit (Sangon) according to the manufacturer’s instructions and used as the template. The 16S rRNA gene was amplified by polymerase chain reaction (PCR) with the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAGCCGCA-3′) (Weisburg et al. 1991). Each 25 μL PCR reaction system consisted of the following reagents: 2 × Taq PCR Mix (Biomiga) 12.5 μL; 10 μM 27F primer 1 μL; 10 μM 1541R primer 1μL; template 1μL; ddH2O 9.5μL. The thermal cycling program was as follows: initial denaturation for 5 min at 95 °C; 30 cycles of denaturation for 30 s at 94 °C; annealing for 60 s at 55 °C; and elongation for 90 s at 72°C, a final extension for 10 min at 72 °C. The PCR product was purified using a UNIQ-10 Column Micro DNA Gel Extraction Kit (Sangon) according to the manufacturer’s instructions. The purified PCR product was sequenced by Biosune Biotechnology Co., Ltd. (Shanghai, China). The obtained gene sequence was compared with sequences in GenBank using BLAST on the National Center for Biotechnology Information (NCBI) website. The related bacterial 16S rRNA gene sequences were downloaded. The sequence similarities between the 16S rRNA genes were calculated using DNAstar software (Burland 2000). ClustalX 2.1 was used for sequence alignment, and the resulting alignment was used for phylogenetic analysis. The average evolutionary divergence over all sequence pairs was computed using maximum composite likelihood model in MEGA 11. The average divergence value for related 16S rRNA genes was 0.05 ± 0.00, confirming that the gene dataset was suitable for generating a neighbor-joining tree. Accordingly, the phylogenetic tree of 16S rRNA sequences was constructed using the neighbor-joining method in MEGA 11 (Tamura et al. 2021).
Morphological and biochemical characterization
Colonies grown on VM agar and Nutrient Agar (NA) (1% peptone, 0.3% Beef extract, 0.5% NaCl, 1.5% agar, pH 7.2 ± 0.2) were observed. Colonies grown on NA were used for Gram staining and observations of bacterial morphology with an optical microscope. The type of Gram staining reaction was also identified by the KOH string method (Powers 1995). Bacterial cells grown on nutrient broth (NB) (1% peptone, 0.3% Beef extract, 0.5% NaCl, pH 7.2 ± 0.2) were used for morphological observation with a scanning electronic microscope (FEI quanta FEG250, USA). A catalase activity test was performed as described previously (Lin et al. 2014). Biochemical characteristics, including oxidase, nitrate reduction, l-tryptophan fermentation, glucose fermentation, hydrolysis of l-arginine, urea, aesculin, gelatin, and 4-nitrophenyl-β-d-galactopyranoside, and assimilation of carbon compounds, were tested using the API 20NE kit (bioMérieux). Other enzyme activities were tested using the API ZYM kit (bioMérieux) according to the manufacturer’s instructions.
Phenolic acid degradation profile
Five phenolic acid media containing, respectively, 0.5 g/L of vanillic acid, ferulic acid, p-coumaric acid, p-hydroxybenzoic acid, or syringic acid as substrate (except for the phenolic acid substrate, the other components were the same as those in the VMY medium) were prepared to detect the phenolic acid substrate profile of strain V4. The V4 culture grown in VMY medium was centrifuged (8000 × g for 10 min at 4 °C), the supernatant was discarded, and the precipitate was suspended in sterilized distilled water to prepare the inoculum (OD600 = 0.6). Then, 10% inoculum (v/v) was added into a sterile phenolic acid medium with vanillic acid, ferulic acid, p-hydroxybenzoic acid, p-coumaric acid, or syringic acid as substrate, respectively. The cultures were incubated at 30 °C and 150 r/min for 5 days, and samples were collected at the beginning and the end of the culture period. After filtration, the substrate content was detected by HPLC (using the same HPLC conditions described above). The substrate degradation rate was calculated as [(initial substrate content − final substrate content)/initial substrate content] × 100% to determine the substrate degradation spectrum of the phenolic acid-degrading bacteria. Each phenolic acid substrate experiment had a control in which 10% (v/v) sterilized distilled water was added instead of strain V4 inoculum. The cultivation condition and substrate detection method of the control were the same as those used for the experiment groups. Each of the experiments was repeated three times.
Influence of environmental conditions on growth of V4
To evaluate the influence of temperature on phenolic acid degradation, 10% (v/v) V4 cultures in VMY medium for each of the different groups were cultured in shaker-incubators at 150 r/min at 20 °C, 30 °C, 37 °C, and 45 °C. Cultures were sampled at 0, 1, and 7 d for measurements of the OD600. To evaluate the influence of pH on phenolic acid degradation, VMY media were prepared, and the pH was adjusted with 5 M NaOH or 5 M HCl to pH values of 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, and 10.0, respectively. 10% (v/v) V4 culture in each medium was incubated at 30°C and 150 r/min, and cultures were sampled at 0, 1, and 7 days for measurement of the OD600. To determine the influence of salinity, different concentrations of NaCl were added to VMY media (as done for the temperature assay, though it had no addition of NaCl). The final concentrations of NaCl were 0%, 0.05%, 1.0%, 2.0%, 3.0%, 4.0%, and 5.0%, respectively. After inoculation with 10% (v/v) V4 culture, each group was incubated at 30 °C and 150 r/min, and cultures were sampled at 0, 1, and 7 days for measurement of the OD600. For evaluating the influence of environmental conditions on the growth of V4, each experiment was conducted with three replicates, and the significant increase (P < 0.05) of OD600 indicated growth of the V4 strain.
Vanillic acid degradation curve and growth curve
For vanillic acid degradation curve determination, 10% (v/v) V4 culture grown in VMY medium was inoculated into a fresh VMY medium. Flasks were cultivated at 30 °C and 150 r/min. Cultures were sampled at 0, 4, 8, 12, 24, 36, and 48 h. Then, the vanillic acid degradation rate was assayed by determining the content with the HPLC technique described above. OD600 at each time was also measured. A control was set up in which 10% (v/v) sterilized distilled water was added in the VMY medium. Experiments were repeated three times.
Whole genome sequencing and analysis
The whole genomic DNA of strain V4T was sequenced by Bei**g Novogene Bioinformatics Technology Co., Ltd. The SMRTbell™ Template Kit (version 1.0) (PacBio, USA) was used to construct a 10-kb SMRTbell library. The NEBNext®Ultra™ DNA Library Prep Kit for Illumina (New England Biolabs, USA) was used to construct a 350-bp library. The library was quantified using a Qubit 4.0 Fluorometer (Invitrogen, Carlsbad, CA, USA), and insert size was estimated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Sequencing was performed using the PacBio Sequel platform and Illumina PE150 platform, according to the respective manufacturer’s instructions. The genome was assembled using SMRT Link 5.0.1 (Ardui et al. 2018; Reiner et al. 2018) (https://www.pacb.com/support/software-downloads/). The GeneMarkS program was used to retrieve the protein-coding genes (Besemer et al. 2001). Transfer RNA (tRNA) genes were predicted by the tRNAscan-SE (Lowe and Eddy 1997). Ribosome RNA (rRNA) genes were analyzed by the rRNAmmer (Lagesen et al. 2007). Genes were annotated using BLAST and the NCBI Non-Redundant Protein Database (NR) (Li et al. 2002). Gene function was also annotated using the Gene Ontology (GO) database, while pathways were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al. 2004, 2006). Proteins were phylogenetically classified using the Clusters of Orthologous Groups (COG) database (Galperin et al. 2015).
Statistical analysis
Statistical analyses were conducted using the SPSS 16.0 software package (SPSS Inc., USA). The degradation rates or OD600 of different groups are presented as means (SD) in text and chart content. Independent-samples t-tests were used to compare the differences between groups in phenolic acid degradation tests. One-way ANOVA was used to analyze the effect of environmental factors on bacterial growth.
Results
Strain isolation and cell morphology
A strain of vanillic acid-degrading bacterium, V4, was isolated on a VM agar plate. V4 degraded 500 mg/L vanillic acid in VM medium with a degradation rate of 100% ± 0% in 5 days, while the degradation rate of the control was − 8.8% ± 2.02%. The difference between the V4 test group and the control group was extremely significant (P < 0.01). V4 formed small, round, smooth, and opaque colonies on the VM agar plate after incubation at 30 °C for 3 days, and the color of colonies changed gradually from milky white to yellow (Fig. 1a). V4 formed small, round, smooth, opaque, and yellow-colored colonies on the NA plate (Fig. 1b) after incubation at 30 °C for 1–2 days. Gram-staining and KOH string tests revealed that V4 was Gram-negative. Scanning electronic microscope observation revealed that V4 was short rod-shaped (0.5–0.6 μm × 0.7–1.2 μm), and the cell morphology is shown in Fig. 1c.
Phylogeny of 16S rRNA gene sequences
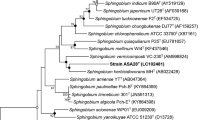
The 16S rRNA gene of strain V4 was sequenced, and the obtained sequence was used as the query for a BLAST search using NCBI. The 16S rRNA gene sequences of representative strains with high similarity to V4 were aligned using ClustalX 2.1. The phylogenetic tree of 16S rRNA genes of V4 and related strains was constructed using MEGA 11 (Fig. 2). The topology of the phylogenetic tree revealed that the V4 strain formed a clade with members of the genus Sphingobium. V4 was most closely related to Sphingobium lactosutens DS20T (= CCM 7540T = MTCC 9471T) (Kumari et al. 2009) (99% sequence similarity). V4 also shared 97.5% sequence similarity with Sphingobium abikonense NBRC 16140T (IAM 12404T = KCTC 2864T) (Kumari et al. 2009). Other species were relatively far from V4 but their sequence similarities with V4 were still over 95%, which indicated that V4 was a member of the genus Sphingobium. Based on 16S rRNA sequence similarities and the phylogenetic analysis, S. lactosutens DS20T and S. abikonense NBRC 16140T were selected as reference strains for comparisons of physiological and biochemical characteristics, and their differential characteristics are summarized in Table 1.
Phylogenetic tree of V4 and related strains based on 16S rRNA gene sequences. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. This analysis involved 19 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1499 positions in the final dataset. Evolutionary analyses were conducted in MEGA11
Biochemical characterization
In the catalase test, V4 tested positive. In the API 20NE tests, V4 was positive for nitrate reduction, L-tryptophan fermentation, aesculin hydrolysis, and 4-nitrophenyl-β-D-galactopyranoside hydrolysis, while negative for glucose fermentation, hydrolysis of L-arginine, urea, and gelatin. Cells assimilated D-glucose, L-arabinose, D-maltose, potassium gluconate, and trisodium citrate, but did not assimilate D-mannose, D-mannitol, N-acetylglucosamine, capric acid, adipic acid, malic acid, or phenylacetic acid. Cytochrome oxidase was positive in V4. In the API ZYM tests, V4 was positive for esterase (C4), esterase lipase (C8), lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase; weakly positive for alkaline phosphatase, naphthol-AS-BI-phosphohydrolase, and β-galactosidase; and negative for leucine arylamidase and acid phosphatase. The detected biochemical characteristics of the V4 strain were similar but different from those of closely related species in Sphingobium (Chaudhary et al. 2017; Kumari et al. 2009); the differentiating characteristics between V4 and the closely related species are summarized in Table 1.
Phenolic acid degradation profile
Five different phenolic acids were tested for substrate degradation. Based on the substrate degradation results, it is clear that V4 degraded vanillic acid, ferulic acid, p-coumaric acid, p-hydroxy benzoic acid, and syringic acid (Fig. 3). After 5 days of culture, degradation rates of the five phenolic acids in the experiment groups containing the V4 strain reached 100 ± 0%, 99.88 ± 0.02%, 99.43 ± 0.19%, 100 ± 0%, and 98.51 ± 0.21%, respectively, while the degradation rates of the control groups were − 1.31 ± 7.82%, 5.53 ± 7.29%, − 2.53 ± 3.93%, − 1.52 ± 8.34%, and 5.52 ± 7.23%, respectively. This indicated that the five phenolic acids tested exhibited no obvious degradation in the control groups, and strain V4 had high degradation ability for the five phenolic acids tested, among which vanillic acid and p-hydroxy benzoic acid were completely degraded. Independent-samples t-tests revealed that there was an extremely significant difference (P < 0.01) between the V4 test group and the control group of each phenolic acid.
At first, we used VM medium to cultivate V4, but we found it grew slowly, besides, V4 cells cultivated in VM medium adhered to the conical flask wall, which brought adverse effect on sampling and subsequent research. So, we added 0.05% yeast extract to improve the growth of V4 in the subsequent study. Accordingly, the subsequent substrate degradation test used VMY medium. Some organic pollutants cannot be used as the only carbon source and energy for microorganisms, and only when there are other organic compounds to provide microbial carbon source or energy can organic pollutants be degraded, the phenomenon was cometabolism. In this study, V4 not only degraded 100% ± 0% of vanillic acid in the VMY medium, but also degraded 100% ± 0% of vanillic acid in the VM medium (without yeast extract). This indicated that the degradation of vanillic acid (a typical phenolic acid) was not the result of cometabolism, but direct degradation.
Influence of environmental conditions on the growth of V4
In the temperature growth test, OD600 measurements showed that V4 grew at temperatures from 20 to 37 °C, with an optimal temperature of 30 °C. One-way ANOVA analysis of OD600 at 1 day revealed there were extremely significant differences (P < 0.01) between 30 °C and other temperature groups. In the pH growth test, OD600 measurements showed that V4 grew at pH values from 5.0 to 9.0, with the optimal pH between 6.0 and 7.0. The difference between the pH 6.0 group and the pH 7.0 group was not significant (P > 0.05). The differences between the pH 6.0 or 7.0 group and the other pH groups were extremely significant (P < 0.01) at 1 day. In the NaCl concentration growth test, OD600 measurements indicated that V4 grew at NaCl concentrations of 0 to 4.0%, with an optimal NaCl concentration of 0 to 0.05% (Fig. 4). The difference between 0% group and 0.05% group was not significant (P > 0.05). The differences between the 0% and 0.05% group and the other NaCl groups were extremely significant (P < 0.01) at 1 day. These results provided reference values for V4 cultivation and future V4 applications in the agricultural environment.
Vanillic acid degradation curve and V4 growth curve
In the VMY medium, both V4 bacterial growth and vanillic acid degradation were detected. The growth curve and vanillic acid degradation curve are shown in Fig. 5. It can be seen that both OD600 and vanillic acid degradation rate increased within 24 h in the V4 test group. At 24 h, the degradation of vanillic acid was 100 ± 0% in the V4 test group; therefore, it is obvious that a stationary stage is reached, while the degradation rate of the control was 3.03 ± 13.06%; the difference between the V4 test group and the control group was extremely significant (P < 0.01). From 8 to 12 h, vanillic acid was degraded most rapidly by the V4 bacterium. During this time, the degradation constant was 11.10%/h; it revealed the degradation efficiency of vanillic acid in V4.
Whole genome sequencing and analysis
To identify candidate genes involved in vanillic acid degradation, we performed whole genome sequencing on Sphingobium sp. V4. The genome assembly indicated that the Sphingobium V4 genome included a circular chromosome (3,209,747 bp) (Fig. 6) and three circular plasmids (1,247,975; 33,206; and 50,153 bp, respectively) (Figs. 7, 8, and 9). The total length was 4,541,081 bp, and the GC content was 64.5%, which fell within the range of 62–67% observed for other members of Sphingobium (Takeuchi et al. 2001).
Chromosome map of Sphingobium sp. V4. From outside to inside: the first layer shows the genome location information; the second layer shows genes encoded on positive and negative strands; the third layer shows the COG functional class of genes on positive and negative strands; the fourth layer shows KEGG functional class of genes on positive and negative strands; the fifth layer shows GO functional class of genes on the positive and negative strand; the sixth layer shows ncRNA; the seventh layer shows the genome G + C mol%; the eighth layer shows the genome GC skew value
The V4 genome contained 4395 coding genes, 59 tRNA genes, and three 5S, three 16S, and three 23S RNA genes. NR annotation was performed on 4200 protein-encoding genes (PEGs), including a VanB gene encoding vanillate O-demethylase oxidoreductase VanB on the chromosome (locus = Chr1:701,288:701,737: -) and a VanA gene encoding vanillate monooxygenase (vanillate O-demethylase oxygenase subunit VanA) on the 1,247,975-bp plasmid (locus = Plas1:504,261:505,313: +), which appeared to be candidate genes for vanillate degradation in strain V4. The VanA amino acid sequence similarities between V4 and Pseudomonas sp. HR199 (Priefert et al. 1997) and between V4 and Pseudomonas resinovorans strain CA10 (Shintani et al. 2013) were 30.9% and 54.9%, respectively. The VanB amino acid sequence similarities between V4 and Pseudomonas sp. HR199 (Priefert et al. 1997) and between V4 and P. resinovorans strain CA10 (Shintani et al. 2013) were 22.8% and 21.5% respectively. COG provided annotations for 3216 genes, and KEGG provided annotations for 4074 genes. GO provided annotations for 2765 entries spanning a total of 44 classifications belonging to molecular function, cellular component, and biological process categories (Fig. 10).
Discussion
Phenolic acids from plant secondary metabolites are key allelopathic compounds that not only affect soil microbial abundance, activity, and community composition, but also induce allelopathy in sensitive plants and thus pose continuous crop** obstacles (Zhalnina et al. 2018; Bai et al. 2019; Zwetsloot et al. 2020). Accordingly, it is important to find and exploit microorganisms that can degrade phenolic acids in order to improve the soil environment and alleviate such continuous crop** obstacles. In recent years, some aromatic- and phenolic acid-degrading Paraburkholderia strains have been used in soil decomposition (Zwetsloot et al. 2020; Wilhelm et al. 2020). Paraburkholderia madseniana is a phenolic acid-degrader isolated from forest soil. It can degrade benzoic acid, phthalic acid, p-hydroxybenzoic acid, and p-coumaric acid (Wilhelm et al. 2020); these properties support the role of P. madseniana in priming the degradation of soil organic matter. In the present study, Sphingobium sp. V4 was revealed to degrade phenolic acids, including vanillic acid, ferulic acid, p-hydroxybenzoic acid, p-coumaric acid, and syringic acid. These capacities suggest Sphingobium sp. V4 is a useful bacterium for phenolic acid degradation of agricultural soil. Additionally, the influence of environmental conditions on V4 growth was also investigated, providing details that could critically guide the application of V4 bacteria to degrade phenol acid under suitable conditions, because efficient growth is associated with efficient phenolic acid degradation. Notably, long-term continuous crop** usually leads to the deterioration of soil physicochemical characteristics, among which soil acidification is an important aspect (Shen et al. 2018). Sphingobium sp. V4 not only grows best at pH 6–7, but also grows well at pH 5. These properties are advantageous for the application of V4 to degrade phenolic acids in acidic soil under continuous crop**.
Sphingobium is a contributor to the catabolism of diverse organic pollutants (Takeuchi et al. 2001). Among the members of Sphingobium, S. abikonense NBRC 16140T, isolated from oil-contaminated soil, can metabolize dibenzothiophene and sulfur-containing organic compounds (Kumari et al. 2009; Anzai et al. 2000; Yamada et al. 1968). Sphingobium naphthae K-3-6T, isolated from oil-contaminated soil, can degrade aliphatic hydrocarbons (Chaudhary et al. 2017). Sphingobium wenxiniae JZ-1T, isolated from activated sludge at a synthetic pyrethroid (SP)-manufacturing wastewater treatment facility, can degrade synthetic pyrethroid pesticides (Wang et al. Wang et al. 2009, Wang et al. 2011). Sphingobium amiense, isolated from river sediment, can degrade nonylphenol (Ushiba et al. 2003). Additionally, Sphingobium sp. SYK-6, isolated from pul** wastewater, is one of the best-characterized degraders of lignin-derived aromatics. SYK-6 can degrade phenolic acids, such as vanillic acid, ferulic acid, and syringic acid (Abe et al. 2005; Masai et al. 2007). Here, Sphingobium sp. V4, isolated from a strawberry field, was determined to degrade a wide variety of phenolic acids (i.e., vanillic acid, ferulic acid, p-hydroxybenzoic acid, p-coumaric acid, and syringic acid). This property indicates that Sphingobium sp. V4 is a promising bacterium for elimination of phenolic acids and agricultural soil improvement.
Vanillic acid is one of the typical phenolic acids. It has strong allelopathy to crops which is involved in continuous crop** obstacles (Wu et al. 2015). So we select vanillic acid as the main compound in this study. The degradation of vanillic acid has been investigated in Pseudomonas spp. The first step in vanillic acid degradation is demethylation, which is catalyzed by a two-component vanillate demethylase (VanA and VanB subunits) that converts vanillic acid to protocatechuic acid. Brunel and Davison characterized the vanA and vanB genes in Pseudomonas, which are located adjacently in the genome (Brunel and Davison 1988). Pseudomonas sp. HR199 (DSM 7063) (Priefert et al. 1997), P. fluorescens BF13 (Civolani et al. 2000), and Acinetobacter have a similar organization of their vanA and vanB genes (Morawski et al. 2000). Nevertheless, Sphingobium sp. SYK-6 degrades vanillic acid by the single component enzyme ligM, which is encoded by the ligM gene (Abe et al. 2005; Masai et al. 2007). In the present study, V4 genomic sequencing revealed a vanB gene on the chromosome and a vanA gene on a large plasmid. Many prokaryotes harbor large plasmids, which are involved in the degradation of recalcitrant organic compounds and in metabolic flexibility (Stolz 2014; Rinke 2022). As mobilome components, plasmids are in constant exchange with more stable chromosomes and contribute to horizontal gene transfer (HGT), an important evolution force among prokaryotes (Koonin and Wolf 2008). Thus, the large plasmid containing vanA and the chromosome containing vanB in the isolated V4 strain are the outcome of prokaryotic evolution, consistent with the genomic diversity of prokaryotes. Other properties of vanillic acid degradation enzymes as well as the genes merit investigation in future research.
Conclusions
The present study isolated the phenolic acid-degrading bacterium V4. Morphological, biochemical, and phylogenetic analysis revealed V4 to be a member of the genus Sphingobium, referred to as Sphingobium sp. V4. Sphingobium sp. V4 effectively degraded vanillic acid, ferulic acid, p-coumaric acid, p-hydroxybenzoic acid, and syringic acid. V4 degraded 500 mg/L vanillic acid completely within 24 h. Sphingobium sp. V4 grew under a wide range of environmental conditions. It not only grew best at pH 6–7, but also grew well at pH 5. These properties are advantageous for the application of V4 in acidic continuous crop** soil. Whole genome analysis revealed that Sphingobium sp. V4 contained one chromosome and three plasmids. Two genes involved in vanillic acid degradation were found in the V4 genome: the gene encoding vanillate O-demethylase oxidoreductase VanB was located on the chromosome, while the gene encoding vanillate monooxygenase (vanillate O-demethylase oxygenase subunit VanA) was located on a large plasmid, illustrating the genomic diversity of prokaryotes. The present study provides support for further investigation and application of the phenolic acid-degrading microorganism to alleviate continuous crop** obstacles in agriculture.
Availability of data and materials
The data generated and analyzed during this study are available in this article. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence and the whole genome sequence of Sphingobium sp. V4 are MZ646136 and CP081001-CP081004 (GCA_029590555.1), respectively.
References
Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M (2005) A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol 187:2030–2037
Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H (2000) Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50:1563–1589
Araki T, Tanatani K, Kamimura N, Otsuka Y, Yamaguchi M, Nakamura M, Masai E (2020) The syringate O-demethylase gene of Sphingobium sp. strain SYK-6 is regulated by DesX, while other vanillate and syringate catabolism genes are regulated by DesR. Appl Environ Microbiol 86:e01712-e1720
Ardui S, Ameur A, Vermeesch JR, Hestand MS (2018) Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res 46:2159–2168
Bai Y, Wang G, Cheng Y, Shi P, Yang C, Yang H, Xu Z (2019) Soil acidification in continuously cropped tobacco alters bacterial community structure and diversity via the accumulation of phenolic acids. Sci Rep 9:12499. https://doi.org/10.1038/s41598-019-48611-5
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Brunel F, Davison J (1988) Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol 170:4924–4930
Burland TG (2000) DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol 132:71–91
Carlsen SC, Kudsk P, Laursen B, Mathiassen SK, Mortensen AG, Fomsgaard IS (2009) Allelochemicals in rye (Secale cereale L.): cultivar and tissue differences in the production of benzoxazinoids and phenolic acids. Nat Prod Commun 4:199–208
Chaudhary DK, Jeong SW, Kim J (2017) Sphingobium naphthae sp. nov., with the ability to degrade aliphatic hydrocarbons, isolated from oil-contaminated soil. Int J Syst Evol Microbiol 67:2986–2993
Chen M, Li X, Yang Q, Chi X, Pan L, Chen N, Yang Z, Wang T, Wang M, Yu S (2014) Dynamic succession of soil bacterial community during continuous crop** of peanut (Arachis hypogaea L.). PLoS One 9:e101355. https://doi.org/10.1371/journal.pone.0101355
Chen S, Yu H, Zhou X, Wu F (2018) Cucumber (Cucumis sativus L.) seedling rhizosphere Trichoderma and Fusarium spp. communities altered by vanillic acid. Front Microbiol 9:1–11
Civolani C, Barghini P, Roncetti A, Ruzzi M, Schiesser A (2000) Bioconversion of ferulic acid into vanillic acid by means of a vanillate- negative mutant of Pseudomonas fluorescens strain BF13. Appl Environ Microbiol 66:2311–2317
Galperin MY, Makarova KS, Wolf YI, Koonin EV (2015) Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261-269
Gulzar A, Siddiqui MB (2015) Root-mediated allelopathic interference of bhringraj (Eclipta alba L.) Hassk. on peanut (Arachis hypogaea) and mung bean (Vigna radiata). Appl Soil Ecol 87:72–80
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354-357
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277-280
Koonin EV, Wolf YI (2008) Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719
Kumari H, Gupta SK, **dal S, Katoch P, Lal R (2009) Sphingobium lactosutens sp. nov., isolated from a hexachlorocyclohexane dump site and Sphingobium abikonense sp. nov., isolated from oil-contaminated soil. Int J Syst Evol Microbiol 59:2291–2296
Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 3:3100–3108
Lin SY, Hameed A, Liu YC, Hsu YH, Lai WA, Huang HI, Young CC (2014) Novosphingobium arabidopsis sp. nov., a DDT-resistant bacterium isolated from the rhizosphere of Arabidopsis thaliana. Int J Syst Evol Microbiol 64:594–598
Liu B, Wan Y, Chen E, Huang M, Chen X, Ni H, He J (2023) Sphingomonas caeni sp. nov., a phenolic acid-degrading bacterium isolated from activated sludge. Antonie Van Leeuwenhoek 116:687–695
Liu J, Li X, Jia Z, Zhang T, Wang X (2017) Effect of benzoic acid on soil microbial communities associated with soil borne peanut diseases. Appl Soil Ecol 11:34–42
Li W, Jaroszewski L, Godzik A (2002) Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18:77–82
Li XG, Zhang TL, Wang XX, Hua K, Zhao L, Han ZM (2013) The composition of root exudates from two different resistant peanut cultivars and their effects on the growth of soil-borne pathogen. Int J Biol Sci 9:164–173
Li X, Lewis EE, Liu Q, Li H, Bai C, Wang Y (2016) Effects of long-term continuous crop** on soil nematode community and soil condition associated with replant problem in strawberry habitat. Sci Rep 6:1–12
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:0955–0964
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotech Bioch 71(1):1–15
Mohan K, Phale PS (2017) Carbon source-dependent inducible metabolism of veratryl alcohol and ferulic acid in Pseudomonas putida CSV86. Appl Environ Microbiol 83(8):e03326-e3416
Morawski B, Segura A, Ornston LN (2000) Substrate range and genetic analysis of Acinetobacter vanillate demethylase. J Bacteriol 182:1383–1389
Powers EM (1995) Efficacy of the Ryu nonstaining KOH technique for rapidly determining gram reactions of food-borne and waterborne bacteria and yeasts. Appl Environ Microbiol 61:3756–3758
Priefert H, Rabenhorst J, Steinbüchel A (1997) Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J Bacteriol 179:2595–2607
Qu XH, Wang JG (2008) Effect of amendments with different phenolic acids on soil microbial biomass, activity, and community diversity. Appl Soil Ecol 39:172–179
Reiner J, Pisani L, Qiao W, Singh R, Yang Y, Shi L, Khan WA, Sebra R, Cohen N, Babu A, Edelmann L, Jabs EW, Scott SA (2018) Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet-Biedl Syndrome 9 (BBS9) deletion. NPJ Genom Med 3: https://doi.org/10.1038/s41525-017-0042-3.
Rinke C (2022) Mystery find of microbial DNA elements called Borgs. Nature 610:635–637
She S, Niu J, Zhang C, ** system. Arch Microbiol 199:267–275
Shen G, Zhang S, Liu X, Jiang Q, Ding W (2018) Soil acidification amendments change the rhizosphere bacterial community of tobacco in a bacterial wilt affected field. Appl Microbiol Biotechnol 102:9781–9791
Shintani M, Hosoyama A, Ohji S, Tsuchikan K, Takarada H, Yamazoe A, Fujita N, Nojiri H (2013) Complete genome sequence of the carbazole degrader Pseudomonas resinovorans strain CA10 (NBRC 106553). Genome Announc 1:e00488-e513. https://doi.org/10.1128/genomeA.00488-13
Stolz A (2014) Degradative plasmids from sphingomonads. FEMS Microbiol Lett 350:9–19
Takeuchi M, Hamana K, Hiraishi A (2001) Proposal of the genus Sphingomonas sensus tricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int J Syst Evol Microbiol 51:1405–1417
Tamura K, Stecher G, Kumar S (2021) MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. https://doi.org/10.1093/molbev/msab120
Tan Y, Cui Y, Li H, Kuang A, Li X, Wei Y, Ji X (2017) Diversity and composition of rhizospheric soil and root endogenous bacteria in Panax notoginseng during continuous crop** practices. J Basic Microbiol 57:337–344
Ushiba Y, Takahara Y, Ohta H (2003) Sphingobium amiense sp. nov., a novel nonylphenol-degrading bacterium isolated from a river sediment. Int J Syst Evol Microbiol 53:2045–2048
Wang B, Guo P, Hang B, Li L, He J, Li S (2009) Cloning of a novel pyrethroid-hydrolyzing carboxylesterase gene from Sphingobium sp. strain JZ-1 and characterization of the gene product. Appl Environ Microbiol 75:5496–5500
Wang B, Guo P, Zheng J, Hang B, Li L, He J, Li S (2011) Sphingobium wenxiniae sp. nov., a synthetic pyrethroid (SP)-degrading bacterium isolated from activated sludge in an SP-manufacturing waste water treatment facility. Int J Syst Evol Microbiol 61:1776–1780
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wilhelm RC, Murphy SJL, Feriancek NM, Karasz DC, DeRito CM, Newman JD, Buckley DH (2020) Paraburkholderia madseniana sp. nov., a phenolic acid-degrading bacterium isolated from acidic forest soil. Int J Syst Evol Microbiol 70:2137–2146
Wu LK, Wang JY, Huang WM, Wu HM, Chen J, Yang YQ, Zhang ZY, Lin WX (2015) Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci Rep 5:15871
**e X, Dai C, Li X, Wu J, Wu Q, Wang X (2017) Reduction of soil-borne pathogen Fusarium solani reproduction in soil enriched with phenolic acids by inoculation of endophytic fungus Phomopsis liquidambari. Biocontrol 62:111–123
Yamada K, Minoda Y, Kodama K, Nakatani S, Akasaki T (1968) Microbial conversion of petro-sulfur compounds. part 1. isolation and identification of dibenzothiophene -utilizing bacteria. Agric Biol Chem 32:840–845
Ye SF, Yu JQ, Peng YH, Zheng JH, Zou LY (2004) Incidence of Fusarium wilt in Cucumis sativus L. is promoted by cinnamic acid, an autotoxin in root exudates. Plant Soil 263:143–150
Yu JQ, Shou SY, Qian YR, Zhu ZJ, Hu WH (2000) Autotoxic potential of cucurbit crops. Plant Soil 223:149–153
Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, Firestone MK, Northen TR, Brodie EL (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480
Zhou X, Wu F (2012) p-Coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f. sp. cucumerinum Owen. PLoS One 7:e48288. https://doi.org/10.1371/journal.pone.0048288
Zhou X, Yu G, Wu F (2012) Soil phenolics in a continuously mono-cropped cucumber (Cucumis sativus L.) system and their effects on cucumber seedling growth and soil microbial communities. Eur J Soil Sci 63:332–340
Zhou X, Zhang J, Pan D, Ge X, ** X, Chen S, Wu F (2018) p-Coumaric can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biol Fert Soils 54:363–372
Zwetsloot MJ, Ucros MJ, Wickings K, Wilhelm RC, Sparks J, Buckley DH, Bauerle TL (2020) Prevalent root-derived phenolics drive shifts in microbial community composition and prime decomposition in forest soil. Soil Biol Biochem 145:107797
Acknowledgements
The authors thank Yongjun Cao and Le Shen for their assistance with laboratory testing.
Funding
This work was supported by the Natural Science Foundation of Shandong Province, China (Grant No. ZR2021MC162).
Author information
Authors and Affiliations
Contributions
C. Z. and Q. M. designed the experiment, analyzed the data, and wrote and revised the manuscript. C. Z., S. L., Q. G., D. L., and Z. L. performed the experiment. H. L., Q. Z., and H. L. assisted in the result analysis and revised the manuscript. Z. D. assisted in the experiment technique, and W. G. and Y. G. conducted part of the experiment.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, C., Liu, S., Guo, Q. et al. Sphingobium sp. V4, a bacterium degrading multiple allelochemical phenolic acids. Ann Microbiol 74, 5 (2024). https://doi.org/10.1186/s13213-024-01750-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13213-024-01750-1