Abstract
Background
Different genotypes of Echinococcus granulosus sensu stricto (s.s.) isolated from livestock and humans have been identified based on cox1 and nad1 genomic fragments. The present study was performed to differentiate the G1/G3 genotypes of Echinococcus granulosus (s.s.) isolated from humans and livestock (sheep and cattle) from Azerbaijan in northwestern Iran, Fars Province in southern Iran, and Van province in Eastern Turkey, using the nad5 gene fragment as a suitable marker to distinguish these two genotypes.
Methods
A total of 60 pathologically confirmed human hydatid cysts and 90 hydatid cyst samples from livestock were collected from Turkey and Iran. PCR was performed on all of the samples, targeting the nad5 gene. Based on PCR product quality, host type, and the geographical area where the samples were obtained, 36 of the samples were sequenced and were used in the phylogenetic analysis.
Results
Out of 36 evaluated samples, 26 (72.2%) samples belonged to G1, and 10 (27.8%) samples belonged to the G3 genotype. Out of 21 samples from Turkey, 16 (76.2%) were G1 and 5 (23.8%) were G3, while out of 15 samples from Iran, 10 (66.7%) were G1 and 5 (33.3%) were the G3 genotype. None of the samples isolated from humans in Iran or from sheep in Turkey were G3. Overall, between the two countries, 18.18% of E. granulosus isolates in cattle, 41.66% of isolates in sheep, and 23.07% of human samples were identified as G3, and the others as the G1 genotype. The G3 genotype was not detected in human samples from Iran or sheep samples from Turkey.
Conclusion
The findings of the study revealed that the G1 genotype of E. granulosus s.s. is the predominant genotype in humans and livestock, both in Turkey and Iran. The ratio of the E. granulosus s.s. G1 to G3 genotype was 3.2 in Turkey and 2 in Iran. The study also further confirmed that the nad5 gene properly differentiated the G1/G3 isolates of E. granulosus from both humans and livestock.
Graphical Abstract

Similar content being viewed by others
Background
Cystic echinococcosis (CE), one of the most important zoonotic diseases, is caused by the larval stage of Echinococcus granulosus sensu lato (s.l.) [1]. E. granulosus s.l. contains a set of different genotypes that differ in life cycle patterns and host types. Currently, 10 genotypes of E. granulosus have been identified using different molecular methods, and are classified into four main groups: sensu stricto (genotypes G1 to G3), equinus (G4), ortleppi (G5), and E. granulosus s.l. genotypes (G6-G10) [2, 3]. The names for genotypes G6–G10, including E. canadensis and E. intermedius, are still under dispute, as recent molecular phylogenetic analysis based on six nuclear genes suggested that E. granulosus s.l. genotypes G6/G7 and G8/G10 can be regarded as two distinct species [4]. The most common hydatid cyst genotypes reported worldwide are two closely related genotypes, G1 and G3, of the sensu stricto strain [5,6,7].
A variety of mitochondrial genes, including cox1, nad1, and atp6 or a fragment sequence of the 12SrRNA gene, as well as the internal transcribed spacer (ITS1) genomic region, have been utilized in various studies for the phylogenetic analysis of E. granulosus [8,9,10,11,12].
Regarding the G1–G3 genotype, a recent study by Kinkar et al. found that G2 is not a valid genotype [13]. Sequences of genomic fragments of cox1 and nad1 have been widely used to differentiate between G1 and G3 genotypes in various studies [14,15,16]. However, given the whole sequences recorded from these genomes, after several decades it has been found that these markers are not able to distinguish between these two genotypes [13]. In a recent study aimed at identifying and differentiating the G1 and G3 genotypes, a simple and practical method was introduced using a 680 bp nad5 genome fragment of E. granulosus s.s. This region contains six useful sites with relatively short lengths that allow us to properly differentiate the G1 and G3 genotypes [13].
Iran and Turkey are considered highly endemic areas for hydatid cysts in both humans and animals [6, 17,18,19,20,21,22,23,24]. In our previous studies on isolates of E. granulosus s.s. from livestock and human populations from Iran and Turkey, different genotypes of the parasite were identified based on cox1 and nad1 genomic fragments [5, 8]. However, since the genomic fragments used in our previous studies were not able to differentiate between the G1 and G3 genotypes, the present study was performed using specific primers and targeting the nad5 genomic fragment to determine the G1/G3 genotype of E. granulosus s.s. isolated from livestock and human samples from Azerbaijan and Fars Provinces in northwestern and southern Iran, and Van Province in eastern Turkey.
Methods
Study area
The study was performed on hydatid cyst samples collected from three regions: Van province from Turkey, located in the east of Van Lake, which is a part of the coldest region in Turkey; East Azerbaijan province as a cold area, located on the mountain range of Iran in the southeast of Urmia Lake; and Fars Province in southern Iran (Table 1). Fars Province has a completely different geographical and climatic condition in comparison with the other two areas in Iran and Turkey, but all three areas have always been considered endemic areas for human CE [5, 8, 24,25,26,27].
Sample preparation
Human and livestock isolated hydatid cysts collected from Turkey and Iran in previous studies, which were identified as G1/G3 genotypes by targeting cox1 and nad1 regions of mitochondrial genome fragments, were re-evaluated by molecular methods. A total of 60 human hydatid cysts, surgically removed and pathologically confirmed, from populations in Tabriz (East Azerbaijan Province, in the northwest of Iran), Shiraz (Fars Province, southern Iran), and Van Province (20 samples from each center), and 90 hydatid cysts from livestock (30 samples from each center) were evaluated in the current study.
DNA extraction
DNA was extracted from each sample as previously described [8]. Briefly, 100 μL of E. granulosus s.s. protoscolices and 25 mg of germinal layers were prepared in different microtubes. A lysis buffer and proteinase K were added to the samples and incubated for 2 h at 60 °C followed by overnight incubation at 37 °C. The rest of the procedure was performed as previously described [8].
Polymerase chain reaction (PCR) and DNA sequencing
Amplification of the nad5 gene was carried out using the two primers EGnd5F1 and EGnd5R1 [28]. For PCR amplification, a final volume of 25 µL reaction containing 3 µL of extracted DNA, 1 µL (10 pm) of each primer, 12.5 µL of 1× Taq DNA Polymerase Master Mix RED (Ampliqon, Odense, Denmark), and 8.5 µL of distilled water (DW) was used. The thermal PCR conditions consisted of initial denaturation at 95 °C for 5 min, and a touchdown protocol with 10 cycles of 95 °C for 20 s, 55 °C for 45 s (annealing temperature progressively reduced by 0.5 °C in each cycle), and 68 °C for 1 min, followed by 25 cycles of 95 °C for 30 s, 50 °C for 45 s, 68 °C for 1 min; finishing with a final elongation step at 68 °C for 5 min. Individual PCR products (2 μL) were examined on 2% agarose gel electrophoresis. PCR was performed on 90 cyst samples (considering the host origin and geographical region where the samples were obtained), targeting the nad5 fragment. Of which, 46 PCR products were selected in terms of the quality of the resulting band on the electrophoresis gel and were sequenced in both directions using the same primers used in the PCR. Out of 46 obtained sequences, the sequences of 36 samples (21 samples from Turkey and 15 samples from Iran, including 11 human samples, 13 sheep, and 12 cattle samples), were of high quality and were used in the phylogenetic analysis.
Phylogenetic and genetic analysis
A total of 36 raw nucleotide sequences were reviewed and analyzed. Consensus sequences were assembled and multiple-aligned with a set of E. granulosus s.s. strains retrieved from the GenBank database. The final aligned sequences with a total of 631 positions were converted in FASTA and MEGA format for further analysis using MEGAX software [29]. The number of base substitutions per site from averaging over all sequence pairs between groups (Kimura 2-parameter genetic distance) was computed using the Kimura 2-parameter model. Codon positions included were 1st + 2nd + 3rd + Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). The best DNA substitution model of HKY+ (- lnL = 1365.6259, k = 89, gamma shape = 0.2 p-inv = 0.6850, AIC = 2909.2517, BIC = 3305.062, kappa = 42.3304 (ti/tv = 15.3337), freq A = 0.1611, freq C = 0.0962, freq G = 0.2563, and freq T = 0.4864) was identified using both the Akaike (AIC) and Bayesian (BIC) information criterion using jModelTest, version0.1.1 [30].
Phylogenetic relationships were reconstructed using a Bayesian inference (BI) tree in MrBayes version 3.1.2, with parameters estimated as part of the analysis. A haplotype network of nad5 data was constructed using the median-joining approach available in PopArt [31]. Genetic diversity based on haplotype diversity (Hd) and nucleotide diversity (π) as well as the number of polymorphic and parsimony-informative sites were measured using DnaSP v5.0 software. The sequences with accession numbers MK682655-56 and MK682657-58 [32] were used in the phylogenetic analysis as references for the G1 and G3 genotypes, respectively. The genotypes G5 (Acc. AB235846) [33], G7 (Acc. MK682636) [32], and G6 (Acc. MH300946) [34] were used as an outgroup for reconstructing the molecular phylogenetic tree.
Results and discussion
The most common genotypes of E. granulosus s.s. causing hydatid cysts in humans and animals are two closely related genotypes, G1 and G3 [16, 35, 36]. For molecular evaluation and phylogenetic analysis of E. granulosus in very close species, different mitochondrial and nuclear genomes have been used. In the present study, the nad5 genomic fragment was used to differentiate between G1 and G3 genotypes in hydatid cysts isolated from humans and livestock in Iran and Turkey.
PCR products of 36 hydatid cyst samples, including 21 samples from Turkey and 15 samples from Iran, were sequenced for the nad5 gene. The sequences were deposited in the GenBank database (NCBI) with related accession numbers. Table 1 shows the sample’s location, the respective host, the parasite genotype, and the assigned accession number in GenBank.
Out of 36 evaluated samples, 26 (72.2%) samples belonged to G1 and 10 (27.8%) samples belonged to the G3 genotype. Out of 21 samples prepared from Turkey, 16 (76.2%) were G1 and 5 (23.8%) were G3, while out of 15 samples prepared from Iran, 10 (66.7%) were G1 and 5 (33.3%) were G3. Overall, in both countries, 18.18% of E. granulosus isolates in cattle, 41.66% of isolates in sheep, and 23.07% of human samples were identified as G3 and others as G1 genotype. Table 2 shows the number of G1/G3 genotypes isolated from different hosts in Iran and Turkey. As the data in Table 2 show, the G3 genotype was not detected in human samples from Iran or sheep samples from Turkey.
Two highly supported major clades were recovered from the BI analysis (Fig. 1). Clade 1 comprises the G1 genotype, infecting humans and livestock from Iran and Turkey, while clade 2 includes the G3 genotype infecting humans and cattle from Turkey and livestock from Iran.
Bayesian phylogenetic tree inferred from 40 nad5 sequences of Echinococcus granulosus sensu stricto (s.s.). Posterior probability values from the Bayesian analysis are indicated at the 100% (**) significance levels. Specimen name includes sample name, host, and country (Table 1)
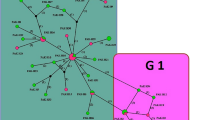
Based on the 631 bp of the nad5 fragment, examined in all 36 sequences, 15 sites were polymorphic, including eight singleton variable sites (two variants) and seven parsimony-informative sites (two variants), resulting in the identification of 14 haplotypes, including 10 haplotypes in G1 and 4 in G3 genotype. Of the 14 haplotypes, 10 distinct haplotypes were observed in the G1 haplogroup from which three were shared between Iranian and Turkish isolates and four were specific to Turkey and three to Iran (Fig. 2). One haplotype (haplotype 3) in the G1 subnetwork with a frequency of 12 was the most frequent haplotype shared between Turkish livestock (3 sheep and 2 cattle), two human samples from Turkey, and four human samples from Iran (Fig. 2). The average number of nucleotide differences and nucleotide divergence between the two main clades G1 and G3 was 6/754 and 1%, respectively. The percentage of the Kimura 2-parameter (K2P) mean genetic distance between G1 and G3 was 1.08%. The percentage of mean K2P distance within the G1 and G3 groups was 0.2% and 0.12%, respectively.
Median-joining haplotype network obtained for 631 bp of mitochondrial nad5 sequences of E. granulosus s.s. Circle size is relative to haplotype frequency; black circles represent extinct or unsampled haplotypes. Hatch marks on the line represent mutational steps between haplotypes. Haplotype colors represent geographic locations of haplotypes as indicated in the right corner of the figure (yellow = Iran, red = Turkey). Host species are indicated with letters inside the haplotypes (C = cattle, S = sheep, H = human). The numbers in parentheses inside haplotypes (in front of letters) indicate the frequency of the haplotype observed in the related host. Haplotype information is summarized in Table 1
Haplotype diversity (Hd) and nucleotide diversity (π) values were 0.769 and 0.002 in G1 sequences, respectively, while these values were 0.533 and 0.0012 in G3 sequences, respectively.
Haplotypes 8, 7, and 5 of nad5 were identified in sheep, cattle, and humans, respectively (Fig. 2). No haplotype in the G1 subnetwork was shared between Iranian and Turkish livestock, but haplotype 3 was shared between sheep and cattle from Turkey (Fig. 2). In the G3 subnetwork, haplotype 11 was shared between Iranian livestock and cattle, and two human samples from Turkey (Fig. 2).
In a study by Romig et al., a low level of differentiation was found between G1 and G3 in the haplotype network, and in a significant proportion of the samples, no distinction was made between the two genotypes [16]. In order to identify and differentiate the G1 and G3 genotypes, Kinkar et al. used data from more than 300 sequences of the parasite's mitochondrial genome to determine the genetic diversity of the two genotypes on a large geographical scale, and introduced a simple new method by sequencing the 680 bp nad5 genome fragment for differentiation of these two genotypes.
Concerning the significance of differentiating G1 and G3, although nuclear gene sequencing data seem to suggest that G1 and G3 form one species, these genotypes have rather different distribution and host range, which provides a good basis to at least suspect some biological relevance. In the phylogeography of many species, including the E. granulosus species complex, even small haplogroups can be of significance in advancing our knowledge. Differentiating G1 and G3 can also potentially help to reveal the geographical origin of the parasite in patients diagnosed with CE.
The present study is a continuation of our previous studies and is designed to highlight the prevalence of Echinococcus genotypes 1 and 3 in Iran and Turkey. The findings of the current study highlight the molecular epidemiology aspect of G1/G3 genotypes in two CE-endemic areas, Iran and Turkey. In addition, with the identification of the G1 and G3 genotypes of Echinococcus, phylogenetic analysis of these two genotypes was documented.
Conclusion
Findings of the current study revealed that the G1 genotype of E. granulosus s.s. is the predominant genotype in humans and livestock in both Turkey and Iran. The ratio of the E. granulosus s.s. G1 to G3 genotype was 3.2 in Turkey and 2 in Iran. The study also further confirms that the nad5 gene properly differentiates the G1/G3 isolates of E. granulosus from both humans and livestock.
Availability of data and materials
Data supporting the conclusions of this article are included within the article. Sequences were deposited in the GenBank database (NCBI) with Accession Numbers MW835719 to 54.
Abbreviations
- CE:
-
Cystic echinococcosis
- ITS1:
-
Internal transcribed spacer
- BI:
-
Bayesian inference
References
Deplazes P, Rinaldi L, Alvarez Rojas CA, Torgerson PR, Harandi MF, Romig T, et al. Global distribution of alveolar and cystic echinococcosis. Adv Parasitol. 2017;95:315–493.
Thompson RCA. Biology and systematics of Echinococcus. Adv Parasitol. 2017;95:65–109.
Thompson RCA, McManus DP. Towards a taxonomic revision of the genus Echinococcus. Trends Parasitol. 2002;18:452–7.
Laurimäe T, Kinkar L, Moks E, Romig T, Omer RA, Casulli A, et al. Molecular phylogeny based on six nuclear genes suggests that Echinococcus granulosus sensu lato genotypes G6/G7 and G8/G10 can be regarded as two distinct species. Parasitology. 2018;145:1929–37.
Barazesh A, Sarkari B, Sarısu G, Hami M, Mikaeili F, Aydın A, et al. Comparative genoty** of Echinococcus granulosus infecting livestock in Turkey and Iran. Turkiye Parazitol Derg. 2019;43:123–9.
Khademvatan S, Majidiani H, Foroutan M, Tappeh KH, Aryamand S, Khalkhali HR. Echinococcus granulosus genotypes in Iran: a systematic review. J Helminthol. 2019;93:131–8.
Rojas CAA, Romig T, Lightowlers MW. Echinococcus granulosus sensu lato genotypes infecting humans–review of current knowledge. Int J Parasitol. 2014;44:9–18.
Barazesh A, Sarkari B, Shahabi S, Halidi AG, Ekici A, Aydemir S, et al. Genetic diversity of Echinococcus granulosus isolated from humans: A comparative study in two cystic echinococcosis endemic areas, Turkey and Iran. Biomed Res Int. 2020;1:3054195.
Rostami Nejad M, Nazemalhosseini Mojarad E, Nochi Z, Fasihi Harandi M, Cheraghipour K, Mowlavi GR, et al. Echinococcus granulosus strain differentiation in Iran based on sequence heterogeneity in the mitochondrial 12S rRNA gene. J Helminthol. 2008;82:343–7.
Rostami Nejad M, Taghipour N, Nochi Z, Mojarad EN, Mohebbi SR, Harandi MF, et al. Molecular identification of animal isolates of Echinococcus granulosus from Iran using four mitochondrial genes. J Helminthol. 2012;86:485–92.
Sarkari B, Mansouri M, Khabisi SA, Mowlavi G. Molecular characterization and seroprevalence of Echinococcus granulosus in wild boars (Sus scrofa) in south-western Iran. Ann Parasitol. 2015;61:269–73.
Yang YR, Rosenzvit MC, Zhang LH, Zhang JZ, McManus DP. Molecular study of Echinococcus in west-central China. Parasitology. 2005;131:547–55.
Kinkar L, Laurimäe T, Sharbatkhori M, Mirhendi H, Kia EB, Ponce-Gordo F, et al. New mitogenome and nuclear evidence on the phylogeny and taxonomy of the highly zoonotic tapeworm Echinococcus granulosus sensu stricto. Infect Genet Evol. 2017;52:52–8.
Busi M, Snábel V, Varcasia A, Garippa G, Perrone V, De Liberato C, et al. Genetic variation within and between G1 and G3 genotypes of Echinococcus granulosus in Italy revealed by multilocus DNA sequencing. Vet Parasitol. 2007;150:75–83.
Yanagida T, Mohammadzadeh T, Kamhawi S, Nakao M, Sadjjadi SM, Hijjawi N, et al. Genetic polymorphisms of Echinococcus granulosus sensu stricto in the Middle East. Parasitol Int. 2012;61:599–603.
Romig T, Ebi D, Wassermann M. Taxonomy and molecular epidemiology of Echinococcus granulosus sensu lato. Vet Parasitol. 2015;213:76–84.
Pestechian N, Hosseini Safa A, Tajedini M, Rostami-Nejad M, Mousavi M, Yousofi H, et al. Genetic diversity of Echinococcus granulosus in center of Iran. Korean J Parasitol. 2014;52:413–8.
Sarkari B, Sadjjadi SM, Beheshtian MM, Aghaee M, Sedaghat F. Human cystic echinococcosis in Yasuj District in Southwest of Iran: an epidemiological study of seroprevalence and surgical cases over a ten-year period. Zoonoses Public Health. 2010;57:146–50.
Sharafi SM, Rostami-Nejad M, Moazeni M, Yousefi M, Saneie B, Hosseini-Safa A, et al. Echinococcus granulosus genotypes in Iran. Gastroenterol Hepatol Bed Bench. 2014;7:82–8.
Altintas N. Past to present: echinococcosis in Turkey. Acta Trop. 2003;85:105–12.
Beyhan YE, Çobanoğlu U, Çelik S, Yılmaz H, Halidi AG. Molecular characterization of human lung and liver cystic echinococcosis isolates in Van Province. Turkey Acta Trop. 2020;206:105451.
Ergin S, Saribas S, Yuksel P, Zengin K, Midilli K, Adas G, et al. Genotypic characterisation of Echinococcus granulosus isolated from human in Turkey. African J Microbiol Res. 2010;4:551–5.
Utuk AE, Simsek S, Koroglu E, McManus DP. Molecular genetic characterization of different isolates of Echinococcus granulosus in east and southeast regions of Turkey. Acta Trop. 2008;107:192–4.
Hosseini G, Sarkari B, Moshfe A, Motazedian MH, Abdolahi KS. Epidemiology of human fascioliasis and intestinal helminthes in rural areas of Boyer-Ahmad Township, southwest Iran; A population based study. Iran J Public Health. 2015;44:1520–5.
Sarkari B, Hosseini F, Abdolahi Khabisi S, Sedaghat F. Seroprevalence of cystic echinococcosis in blood donors in Fars province, southern Iran. Parasite Epidemiol Control. 2016;15:8–12.
Shahriarirad R, Erfani A, Eskandarisani M, Rastegarian M, Sarkari B. Uncommon locations of cystic echinococcosis: a report of 46 cases from southern Iran. Surg Res Pract. 2020;1:2061045.
Shahriarirad R, Erfani A, Eskandarisani M, Rastegarian M, Taghizadeh H, Sarkari B. Human cystic echinococcosis in southwest Iran: a 15-year retrospective epidemiological study of hospitalized cases. Trop Med Health. 2020;19(48):49.
Kinkar L, Laurimäe T, Acosta-Jamett G, Andresiuk V, Balkaya I, Casulli A, et al. Distinguishing Echinococcus granulosus sensu stricto genotypes G1 and G3 with confidence: a practical guide. Infect Genet Evol. 2018;64:178–84.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6.
Leigh JW, Bryant D. Popart: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–6.
Laurimäe T, Kinkar L, Varcasia A, Dessì G, Sgroi G, D’Alessio N, et al. First detection of zoonotic tapeworm Echinococcus granulosus sensu lato genotype G7 in continental Italy. Parasitol Res. 2019;118:2193–201.
Nakao M, McManus D, Schantz P, Craig P, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology. 2006;134:713–22.
Laurimäe T, Kinkar L, Romig T, Omer RA, Casulli A, Umhang G, et al. The benefits of analysing complete mitochondrial genomes: deep insights into the phylogeny and population structure of Echinococcus granulosus sensu lato genotypes G6 and G7. Infect Genet Evol. 2018;64:85–94.
Alvarez Rojas CA, Romig T, Lightowlers MW. Echinococcus granulosus sensu lato genotypes infecting humans–review of current knowledge. Int J Parasitol. 2014;44:9–18.
Harandi MF, Hobbs RP, Adams PJ, Mobedi I, Morgan-Ryan UM, Thompson RCA. Molecular and morphological characterization of Echinococcus granulosus of human and animal origin in Iran. Parasitology. 2002;125:367–73.
Funding
The study was financially supported by the office of Vice-Chancellor for Research of Shiraz University of Medical Sciences (Grant No. 97–01-106–17955).
Author information
Authors and Affiliations
Contributions
BS and SS were involved in the study design. SS and AB were involved in performing the experiments and data collection. BS, SS, and AB were involved in the data analysis and preparation of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Research Ethics Committee of Shiraz University of Medical Sciences (SUMS, Iran).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shahabi, S., Sarkari, B. & Barazesh, A. Echinococcus granulosus sensu stricto G1 is the predominant genotype in human and livestock isolates from Turkey and Iran, based on mitochondrial nad5 gene differentiation. Parasites Vectors 14, 369 (2021). https://doi.org/10.1186/s13071-021-04869-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-04869-1