Abstract
Background
The morphology of the adrenal tumor and the clinical statistics of the adrenal tumor area are two crucial diagnostic and differential diagnostic features, indicating precise tumor segmentation is essential. Therefore, we build a CT image segmentation method based on an encoder–decoder structure combined with a Transformer for volumetric segmentation of adrenal tumors.
Methods
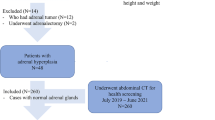
This study included a total of 182 patients with adrenal metastases, and an adrenal tumor volumetric segmentation method combining encoder–decoder structure and Transformer was constructed. The Dice Score coefficient (DSC), Hausdorff distance, Intersection over union (IOU), Average surface distance (ASD) and Mean average error (MAE) were calculated to evaluate the performance of the segmentation method.
Results
Analyses were made among our proposed method and other CNN-based and transformer-based methods. The results showed excellent segmentation performance, with a mean DSC of 0.858, a mean Hausdorff distance of 10.996, a mean IOU of 0.814, a mean MAE of 0.0005, and a mean ASD of 0.509. The boxplot of all test samples' segmentation performance implies that the proposed method has the lowest skewness and the highest average prediction performance.
Conclusions
Our proposed method can directly generate 3D lesion maps and showed excellent segmentation performance. The comparison of segmentation metrics and visualization results showed that our proposed method performed very well in the segmentation.
Similar content being viewed by others
Background
The adrenal glands are important secretory organs that are positioned above the kidneys on both sides of the body. The adrenal glands can be divided into cortical and medullary layers, which mainly secrete adrenal corticosteroids, epinephrine, and norepinephrine, and play an important role in maintaining the normal functioning of body organs [1, 2]. Adrenal tumors are formed by abnormal proliferation of local adrenal tissue cells and are pathologically classified into benign tumors (expansive growth) and malignant tumors (invasive growth) according to their biological behavior [3,4,5]. Both benign and malignant tumor types can affect hormone secretion and lead to hypertension, hyperglycemia, and cardiovascular diseases [6,7,8].
Computed tomography (CT) is an effective tool for diagnosing adrenal tumors, because it is non-invasive, produces clear images, and has high diagnostic efficiency. Because the morphology of the tumor (mainly refers to the smoothness of tumor edge) and the clinical statistics (mainly refers to the difference in density and gray level within the tumor) of the tumor area are two crucial diagnostic and differential diagnostic features, precise segmentation of the tumor is essential. There have been many studies evaluating traditional methods for adrenal tumor segmentation, such as classifying CT images pixel-by-pixel with a random forest classifier [9], and applying a localized region-based level set method (LRLSM) to segment the region of interest (ROI) [10]. Some studies have proposed segmentation frameworks that combine various algorithms to obtain better segmentation performance [11,12,13]. These include the K-means singular value factorization (KSVD) algorithm, region growing (RG), K-means clustering, and image erosion.
In recent years, with the continuous improvement in computing power and the development of deep learning techniques [14,30] is a relatively mature application, and segmentation of brain tumors [31, 32], kidney [33], kidney tumors [34, \(\frac{\mathrm{W}}{16}\times \frac{\mathrm{H}}{16}\times \frac{\mathrm{D}}{16}\). Instead of directly segmenting the original image into patches, the Transformer can model local contextual features in the spatial and depth dimensions of the high-level feature map. The Transformer requires the input to be sequence data, so we use the Reshape operation to flatten the feature map to \(256 \times \mathrm{N}\), where \(\mathrm{N}=\frac{\mathrm{W}}{16}\times \frac{\mathrm{H}}{16}\times \frac{\mathrm{D}}{16}\).
The importance of the position of each pixel in the image cannot be ignored, and such spatial information is indispensable for the accurate segmentation of tumors. Therefore, we also add learnable position embedding to reconstruct location information.
(2) Transformer layer
The architecture of the Transformer layer, which consists of a multi-head attention (MHA) and feed forward network, is shown in Fig. 1.
The MHA consists of eight single attention heads, which can be viewed as map** a collection of query vectors to output vectors based on key-value pairs. The details are shown in Formulas 1 and 2.
where \({\mathrm{W}}_{\mathrm{i}}^{\mathrm{O}}\in {\mathrm{R}}^{8{\mathrm{d}}_{\mathrm{v}}\times \mathrm{D}}\), \({\mathrm{W}}_{\mathrm{i}}^{\mathrm{Q}}\in {\mathrm{R}}^{\mathrm{D}\times {\mathrm{d}}_{\mathrm{k}}}\) and \({\mathrm{W}}_{\mathrm{i}}^{\mathrm{V}}\in {\mathrm{R}}^{\mathrm{D}\times {\mathrm{d}}_{\mathrm{v}}}\) are learnable parameter matrices, and Q, K, and V are query, key, and value, respectively.
The Feed Forward Network comprises a fully connected neural network and an activation function.
Network decoder
Before up-sampling, patches need to be mapped (feature map**) to the original space, then the up-sampling operation is performed. The decoder also up-samples four times, with the overall operation corresponding to the down-sampling. Because some spatial context information is inevitably lost during down-sampling, we use a skip connection to connect the feature maps corresponding to the down-sampling stage. This skip connection ensures that the new feature maps contain both shallow low-level information and high-level abstract semantic information.
Comparison details
To verify the effectiveness of our proposed method, we make a comparison with the mainstream medical image segmentation methods. The implementation details of the compared methods are as follows:
-
a.
3D U-Net first constructs a \(7\times 7\) convolution block, then constructs four encoder and decoder blocks. Finally, a final convolution block is constructed, including a transposed convolution and two sub-convolution blocks.
-
b.
TransBTS is constructed with a series of components. It starts with four encoder blocks, which are then followed by a classical Transformer module containing four Transformer layers, each equipped with eight heads of attention. Subsequently, four decoder blocks are added to the model. To complete the architecture, TransBTS finally incorporates a convolutional layer and utilizes the softmax function.
-
c.
ResUNet combines ResNet and U-Net by integrating the residual block in each encoder and decoder block. During skip-connection phase, convolutional blocks are additionally constructed to match the dimensions of the encoder output and the decoder output at the corresponding stage.
-
d.
UNet++ aggregates 1 to 4 layers of U-Net together and builds a convolutional layer and sigmoid function at the end.
-
e.
The structure of Attention U-Net is generally the same as that of U-Net, with the difference that Attention U-Net adds a layer of attention gates before skip-connection.
-
f.
Channel U-Net builds six encoder and decoder blocks and adds the Global Attention Upsample module before skip-connection.
Training details
All networks were implemented based on the PyTorch framework, and four NVIDIA RTX 3080 with 10 GB memory were used for training. We divided the entire data set into 80% training set and 20% testing set. The testing set is finally used to test the segmentation performance of our proposed model, and the results can be seen in Fig. 1. Given the large size of a single sample, 32 consecutive slices are randomly selected from a sample as input data. We adopted the Adam optimizer in the training process. The weight decay was set to \(1\times {10}^{-5}\), the learning rate was \(2\times {10}^{-4}\) and \(4\times {10}^{-7}\) when the epoch, respectively, reached 0 and 999 (all networks were trained for 1000 epochs), the batch size was set to 2, and the random number seed was set to 1000.
Evaluation metrics
To evaluate the effectiveness of our proposed method, we used the Dice coefficient (DSC) and Intersection over union (IOU), which are widely used to evaluate the similarity between segmentation results and ground truth data in medical image segmentation. Furthermore, we used the Hausdorff distance and Average surface distance (ASD) to evaluate the similarity of the surface between the segmentation results and ground truth. Mean average error (MAE) is used to assess the absolute error. These metrics are defined in Eqs. 3, 4, 5, 6, and 7, respectively:
For statistical analysis, we compare the difference between the prediction results of the proposed method and other methods. We first use the Levene test to check the homogeneity of variance and then conduct Student's t test between our proposed method and other methods.
Availability of data and materials
The data used in this study are available from the corresponding author on reasonable request.
Abbreviations
- DSC:
-
Dice coefficient
- IOU:
-
Intersection over union
- CT:
-
Computed tomography
- LRLSM:
-
Localized region-based level set method
- ROI:
-
Region of interest
- KSVD:
-
K-means singular value factorization
- RG:
-
Region growing
- KaCNN:
-
Knowledge-assisted convolutional neural network
- CNN:
-
Convolutional neural network
- DSConv:
-
Deep separable convolution
- MHA:
-
Multi-head attention
- NLP:
-
Natural language processing
- ASD:
-
Average surface distance
- MAE:
-
Mean average error
References
Lyraki R, Schedl A. Adrenal cortex renewal in health and disease. Nat Rev Endocrinol. 2021;17(7):421–34.
Capellino S, Claus M, Watzl C. Regulation of natural killer cell activity by glucocorticoids, serotonin, dopamine, and epinephrine. Cell Mol Immunol. 2020;17(7):705–11.
Zhu F, Zhu X, Shi H, Liu C, Xu Z, Shao M, Tian F, Wang J. Adrenal metastases: early biphasic contrast-enhanced CT findings with emphasis on differentiation from lipid-poor adrenal adenomas. Clin Radiol. 2021;76(4):294–301.
Athanasouli F, Georgiopoulos G, Asonitis N, Petychaki F, Savelli A, Panou E, Angelousi A. Nonfunctional adrenal adenomas and impaired glucose metabolism: a systematic review and meta-analysis. Endocr. 2021;74(1):50–60.
Chen WC, Baal JD, Baal U, Pai J, Gottschalk A, Boreta L, Braunstein SE, Raleigh DR. Stereotactic body radiation therapy of adrenal metastases: a pooled meta-analysis and systematic review of 39 studies with 1006 patients. Int J Radiat Oncol Biol Phys. 2020;107(1):48–61.
Grasso M, Boscaro M, Scaroni C, Ceccato F. Secondary arterial hypertension: from routine clinical practice to evidence in patients with adrenal tumor. High Blood Press Cardiovasc Prev. 2018;25(4):345–54.
Szychlińska M, Baranowska-Jurkun A, Matuszewski W, Wołos-Kłosowicz K, Bandurska-Stankiewicz E. Markers of subclinical cardiovascular disease in patients with adrenal incidentaloma. Medicina. 2020;56(2):69.
Wrenn SM, Pandian T, Gartland RM, Fong ZV, Nehs MA. Diabetes mellitus and hyperglycemia are associated with inferior oncologic outcomes in adrenocortical carcinoma. Langenbecks Arch Surg. 2021;406(5):1599–606.
Saiprasad G, Chang C-I, Safdar N, Saenz N, Siegel E. Adrenal gland abnormality detection using random forest classification. J Digit Imaging. 2013;26(5):891–7.
Tang S, Guo Y, Wang Y, Cao W, Sun F, editors. Segmentation and 3D visualization of pheochromocytoma in contrast-enhanced CT images. 2014 International Conference on Audio, Language and Image Processing; 2014: IEEE.
Chai HC, Guo Y, Wang YY. Automatic Segmentation of Adrenal Tumor in CT Images Based on Sparse Representation. J Med Imaging Health Inform. 2015;5(8):1737–41. https://doi.org/10.1166/jmihi.2015.1637.
Koyuncu H, Ceylan R, Erdogan H, Sivri M. A novel pipeline for adrenal tumour segmentation. Comput Methods Programs Biomed. 2018;159:77–86. https://doi.org/10.1016/j.cmpb.2018.01.032.
Barstugan M, Ceylan R, Asoglu S, Cebeci H, Koplay M. Adrenal tumor segmentation method for MR images. Comput Methods Programs Biomed. 2018;164:87–100. https://doi.org/10.1016/j.cmpb.2018.07.009.
Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7. https://doi.org/10.1126/science.1127647.
Kaiming H, **angyu Z, Shaoqing R, Jian S. Deep residual learning for image recognition. IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016;2016:770–8. https://doi.org/10.1109/cvpr.2016.90.
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative Adversarial Networks. Commun ACM. 2020;63(11):139–44. https://doi.org/10.1145/3422622.
Wang W, Chen C, Ding M, Yu H, Zha S, Li J, editors. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. Med Image Comput Comput Assist Interv (MICCAI); 2021 2021. Sep 27-Oct 01; Electr Network2021. https://doi.org/10.1007/978-3-030-87193-2_11.
Lei T, Wang R, Zhang Y, Wan Y, Liu C, Nandi AK. DefED-Net: Deformable encoder-decoder network for liver and liver tumor segmentation. IEEE Trans Radiat Plasma Med Sci. 2021;6(1):68–78.
Cicek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. Med Image Comput Comput Assist Interv. 2016: 424–32 p. https://doi.org/10.1007/978-3-319-46723-8_49.
Zhao Y, Li H, Wan S, Sekuboyina A, Hu X, Tetteh G, Piraud M, Menze B. Knowledge-aided convolutional neural network for small organ segmentation. IEEE J Biomed Health Inform. 2019;23(4):1363–73. https://doi.org/10.1109/jbhi.2019.2891526.
Bi L, Kim J, Su T, Fulham M, Feng D, Ning G, Ieee, editors. Adrenal Lesions Detection on Low-Contrast CT Images using Fully Convolutional Networks with Multi-Scale Integration. IEEE 14th International Symposium on Biomedical Imaging (ISBI) - From Nano to Macro; 2017 2017. Apr 18–21; Melbourne, AUSTRALIA2017.
Alimu P, Fang C, Han Y, Dai J, **e C, Wang J, Mao Y, Chen Y, Yao L, Lv C, Xu D, **e G, Sun F. Artificial intelligence with a deep learning network for the quantification and distinction of functional adrenal tumors based on contrast-enhanced CT images. Quant Imaging Med Surg. 2023;13(4):2675–87. https://doi.org/10.21037/qims-22-539.
Jha D, Smedsrud PH, Johansen D, de Lange T, Johansen HD, Halvorsen P, Riegler MA. A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation. IEEE J Biomed Health Inform. 2021;25(6):2029–40.
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. Unet++: A nested u-net architecture for medical image segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support: Springer; 2018. p. 3–11.
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B. Attention u-net: Learning where to look for the pancreas. ar**v preprint ar**v:180403999. 2018.
Chen Y, Wang K, Liao X, Qian Y, Wang Q, Yuan Z, Heng P-A. Channel-Unet: a spatial channel-wise convolutional neural network for liver and tumors segmentation. Front Genet. 2019;10:1110.
Alsallakh B, Kokhlikyan N, Miglani V, Yuan J, Reblitz-Richardson O. Mind the Pad--CNNs can Develop Blind Spots. ar**v preprint ar**v:201002178. 2020.
Shi J, Ye Y, Zhu D, Su L, Huang Y, Huang J. Comparative analysis of pulmonary nodules segmentation using multiscale residual U-Net and fuzzy C-means clustering. Comput Methods Programs Biomed. 2021;209:106332.
Ni Y, **e Z, Zheng D, Yang Y, Wang W. Two-stage multitask U-Net construction for pulmonary nodule segmentation and malignancy risk prediction. Quant Imaging Med Surg. 2022;12(1):292.
Zhou Z, Gou F, Tan Y, Wu J. A cascaded multi-stage framework for automatic detection and segmentation of pulmonary nodules in develo** countries. IEEE J Biomed Health Inform. 2022.
Parmar B, Parikh M, editors. Brain tumor segmentation and survival prediction using patch based modified 3D U-Net. International MICCAI Brainlesion Workshop; 2020. Springer.
Zhang D, Huang G, Zhang Q, Han J, Han J, Yu Y. Cross-modality deep feature learning for brain tumor segmentation. Pattern Recogn. 2021;110: 107562.
da Cruz LB, Araújo JDL, Ferreira JL, Diniz JOB, Silva AC, de Almeida JDS, de Paiva AC, Gattass M. Kidney segmentation from computed tomography images using deep neural network. Comput Biol Med. 2020;123: 103906.
da Cruz LB, Júnior DAD, Diniz JOB, Silva AC, de Almeida JDS, de Paiva AC, Gattass M. Kidney tumor segmentation from computed tomography images using DeepLabv3+ 2.5 D model. Expert Syst Appl. 2022;192:116270.
Hou X, **e C, Li F, Wang J, Lv C, **e G, Nan Y, editors. A triple-stage self-guided network for kidney tumor segmentation. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI); 2020: IEEE.
Le DC, Chinnasarn K, Chansangrat J, Keeratibharat N, Horkaew P. Semi-automatic liver segmentation based on probabilistic models and anatomical constraints. Sci Rep. 2021;11(1):1–19.
Winkel DJ, Weikert TJ, Breit H-C, Chabin G, Gibson E, Heye TJ, Comaniciu D, Boll DT. Validation of a fully automated liver segmentation algorithm using multi-scale deep reinforcement learning and comparison versus manual segmentation. Eur J Radiol. 2020;126: 108918.
Zhao J, Li D, **ao X, Accorsi F, Marshall H, Cossetto T, Kim D, McCarthy D, Dawson C, Knezevic S. United adversarial learning for liver tumor segmentation and detection of multi-modality non-contrast MRI. Med Image Anal. 2021;73: 102154.
Luo G, Yang Q, Chen T, Zheng T, **e W, Sun H. An optimized two-stage cascaded deep neural network for adrenal segmentation on CT images. Comput Biol Med. 2021. https://doi.org/10.1016/j.compbiomed.2021.104749.
Acknowledgements
Not applicable.
Funding
This research was supported by Natural Science Foundation of Zhejiang Province under Grant Nos. LQ20F020014 and JWZ22E090001.
Author information
Authors and Affiliations
Contributions
LPW, MTY, YJL, QCQ, ZFN, HFS and JW conceived and designed the study. LPW, MY, and JW contributed to the literature search. JW, HFS, ZFN and YJL contributed to data collection. MTY, JW, and YJL contributed to data analysis. MTY, JW and WLP contributed to data interpretation. MTY and LPW contributed to the figures. MTY, LPW and JW contributed to writing of the report.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This retrospective study was approved by the ethics committee of Tongde Hospital of Zhejiang Province (Approval Number 2022-183) and conducted in accordance with the tenets of the Declaration of Helsinki. The data were anonymized, and the requirement for informed consent was waived.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, L., Ye, M., Lu, Y. et al. A combined encoder–transformer–decoder network for volumetric segmentation of adrenal tumors. BioMed Eng OnLine 22, 106 (2023). https://doi.org/10.1186/s12938-023-01160-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12938-023-01160-5