Abstract
Background
Diarrhea is one of the most common diseases in pig industry, which seriously threatens the health of piglets and causes huge economic losses. Enterotoxigenic Escherichia coli (ETEC) F4 is regarded as the most important cause of diarrhea in piglets. Some pigs are naturally resistant to those diarrheas caused by ETEC-F4, because they have no F4 receptors (F4R) on their small intestine epithelial cells that allow F4 fimbriae adhesion. Circular RNA (circRNA) has been shown to play an important regulatory role in the pathogenesis of disease. We hypothesized that circRNAs may also regulate the adhesion of piglet small intestinal epithelial cells to ETEC F4 fimbriae. However, the circRNA expression profiles of piglets with different Enterotoxigenic Escherichia coli F4 fimbriae (ETEC-F4ac) adhesion phenotypes are still unclear, and the intermediate regulatory mechanisms need to be explored. Hence, the present study assessed the circRNA expression profiling in small intestine epithelial cells of eight male piglets with different ETEC-F4 adhesion phenotypes and ITGB5 genotypes to unravel their regulatory function in susceptibility to ETEC-F4ac diarrhea. Piglets were divided into two groups: non-adhesive group (n = 4) with CC genotype and adhesive group (n = 4) with TT genotype.
Results
The RNA-seq data analysis identified 13,199 circRNAs from eight samples, most of which were exon-derived. In the small intestine epithelial cells, 305 were differentially expressed (DE) circRNAs between the adhesive and non-adhesive groups; of which 46 circRNAs were upregulated, and 259 were downregulated. Gene ontology and KEGG enrichment analysis revealed that most significantly enriched DE circRNAs’ host genes were linked to cytoskeletal components, protein phosphorylation, cell adhesion, ion transport and pathways (such as adherens junction, gap junction) associated with ETEC diarrhea. The circRNA-miRNA-mRNA interaction network was also constructed to elucidate their underlying regulatory relationships. Our results identified several candidate circRNAs that affects susceptibility to ETEC diarrhea. Among them, circ-SORBS1 can adsorb ssc-miR-345-3p to regulate the expression of its host gene SORBS1, thus improving cell adhesion.
Conclusion
Our results provided insights into the regulation function of circRNAs in susceptibility to ETEC diarrhea of piglets, and enhanced our understanding of the role of circRNAs in regulating ETEC diarrhea, and reveal the great potential of circRNA as a diagnostic marker for susceptibility of ETEC diarrhea in piglets.
Similar content being viewed by others
Background
Diarrhea is the most common disease in pig industry, which seriously endangers the health of piglets and the stable development of pig industry. Enterotoxigenic Escherichia coli (ETEC) expressing the F4 fimbriae is a major cause of diarrhea in neonatal and pre-weaned piglets [1], which leads to considerable economical loss in the pig industry. Three antigenic variants of F4 have been described: F4ab, F4ac and F4ad, of which F4ac is the most prevalent [2]. Studies have shown that the resistance and susceptibility phenotype of piglets to E. coli diarrhea is determined by the presence or absence of F4 receptors on their small intestinal epithelial cells [3]. In our previous study, ITGB5 was found to be a key gene related to the adhesion phenotypes (Small intestinal epithelial cells with or without F4 receptors) [4]. ITGB5 genoty** can effectively distinguish piglets susceptible or resistant to E. coli diarrhea [5]. The small intestinal epithelial cells of piglets with CC genotype were non-adhesive and resistant to ETEC-F4ac diarrhea, whereas piglets with TT genotype were adhesive and susceptible to ETEC-F4ac diarrhea.
Circular RNA (circRNA), a novel class of non-coding RNA, is characterized by a closed-loop structure generated by pre-mRNA back splicing [6]. Different from the traditional linear RNA (including 5 'and 3' tail), circRNA molecules have a closed ring structure and are not affected by RNA exonuclease, thus their expression is more stable and not easy to degrade [7]. Using high-throughput RNA sequencing (RNA-seq) techniques, recent results have shown that a large number of circRNAs are endogenous, stable and widely expressed in mammalian cells, often exhibiting cell type-specific, tissue-specific or developmental stage-specific expression [8,9,10]. In terms of function, recent studies have shown that circRNA molecules are rich in microRNA (miRNA) binding sites and act as miRNA sponges in cells [11], thereby relieving the inhibition of miRNA on target genes and increasing the expression level of target genes. This mechanism of action is known as the competitive endogenous RNA (ceRNA) mechanism. Li et al. [12] found that a novel circ-PPARA could promote the formation of intramuscular fat in pigs through the adsorption of miR-429 and miR-200b. In addition, circRNA are involved in the development of various disease conditions, such as cardiovascular diseases [13], diabetes [14], neurological diseases [15] and cancer [16]. Emerging evidence suggests that circRNAs may be potential new clinical diagnostic markers or therapeutic approaches for many diseases [17]. Yan et al. [18] investigated circRNA expression profiles in spleen of piglets infected with Clostridium perfringens type C, identifying eight circRNAs associated with necrotizing enteritis caused by Clostridium perfringens type C. Chen et al. [19] analyzed the circRNA expression profile during porcine endemic diarrhea virus (PEDV) infection in IPEC-J2 cell line and identified 26,670 candidate circRNAs.
In this study, we comprehensively analyzed characteristics of circRNA in the adhesive and non-adhesive small intestinal epithelial cells of piglets using RNA-seq data and bioinformatics methods, and explored the role of DE circRNAs in susceptibility to ETEC-F4ac. We also constructed ceRNA network including circRNA, miRNA and mRNA to identify molecule markers involved in susceptibility and resistance to ETEC-F4ac Diarrhea. The results of this study enhance our understanding of the role of circRNAs in regulating ETEC diarrhea resistance, and reveal the great potential of circRNA as a therapeutic target to biological treatment for ETEC diarrhea in piglets.
Results
Characterization of circRNAs in porcine small intestinal epithelial cells
A summary of RNA-seq data from eight porcine small intestinal epithelial cell samples is shown in Table 1. Two kinds of software (find_circ and CIRI2) were used for identification of circRNAs based on back-spliced reads produced from high-throughput RNA sequencing data. We identified 13,199 circRNAs with at least two independent back-spliced junction reads via two kinds of software (Additional file 1: Table S1). The obtained 13,199 circRNAs were aligned with circNet [20] and circAtlas [21] databases, among which 12,020 circRNAs (91%) were known, and 1179 circRNAs were novel (Fig. 1a). Chromosome distribution of those circRNAs is concentrated on chromosome 1, and the least on chromosome Y. Some circRNAs come from mitochondria, and the rest of the chromosomes are roughly evenly distributed (Fig. 1b). Most of the circRNAs identified were exonic circRNA (12,079, accounting for about 91.5%), and 796 circRNAs were intronic (Fig. 1e). These circRNAs were generated from 4400 genes, among which 42.95% genes produced only one circRNA isoform (Fig. 1c). The remaining 324 circRNAs originated from intergenic regions. The most circRNAs were made up of 2–3 exons, and few are more than 10 exons (Fig. 1d). The counts of the back-spliced junction reads of 8 samples were normalized as the spliced reads per billion map** (SRPBM) (Additional file 1: Table S2). The circRNAs originated from exon, intron and intergenic region showed no significant changes in expression abundance (SRPBM) (Fig. 1f). Then we aligned the flanking intron pairs of exonic circRNAs to identify reverse complementary matches (RCMs) using Basic Local Alignment Search Tool (BLAST). Among the identified RCMs, Short interspersed nuclear element (SINE) and Simple_repeat accounted for 77.5% and 12.7%, respectively (Fig. 1g).
Profiling of circRNA in small intestinal epithelial cells of piglets. a CircRNAs identified in piglet intestinal epithelial cells (IEC) were overlapped with circRNAs annotated in circNet and circAtlas databases. b The distribution of identified circRNAs in different chromosomes. c The distribution of host genes encoding different number of circRNAs in piglet intestinal epithelial cells. d The distribution of number of exons that form the exonic circRNAs. e The proportion of different categories of circRNAs in intestinal epithelial cells of piglets in each sample. f Cumulative distribution of different categories of circRNA expression. g The proportion of different categories of RCMs in the flanking introns of exonic circRNAs. IEC: intestinal epithelial cells; SRPBM: spliced reads per billion map**; LINE: long interspersed nuclear elements; LTR: long terminal repeat; SINE: short interspersed nuclear element
Differentially expression circRNAs in piglet small intestinal epithelial cells that susceptible/resistant to diarrhea
We further compared the circRNAs in porcine intestinal epithelial cells of the adhesive and non-adhesive groups to identify molecular markers associated with E. coli diarrhea. No significant difference was observed in the distribution density of circRNA expression determined based on the SRPBM value obtained from the RNA-seq data of the two groups (Fig. 2a). Next, 305 differentially expressed circRNAs (DEC) were obtained by using DEseq2 R package between adhesive and non-adhesive group (Additional file 1: Table S3), of which 259 were down-regulated and 46 were up-regulated in the non-adhesive group (Fig. 2b). The clustering heatmap comparison showed some circRNAs predominately expressed in adhesive group and some mainly expressed in non-adhesive group (Fig. 2c).
DE circRNAs in porcine small intestine epithelial cell with adhesive and non-adhesive group. a Density curves of circRNA expression levels in different samples. b Volcano plots depicting DE circRNAs in adhesive group and non-adhesive group. c Heatmap showing the expression of all DE circRNAs identified in this research
Functional enrichment analysis of differentially expressed circRNAs revealed their regulatory role in the biogenesis of piglets E. coli diarrhea
The present study performed GO and KEGG analyses of the DE circRNA host genes to elucidate their roles in ETEC-F4ac diarrhea. Functional annotation identified 171 significantly enriched GO terms and 46 significantly enriched pathways (Additional file 1: Table S4). The significantly GO terms of DE circRNA host genes were preferentially enriched in functions related to cytoskeletal components, cell junction, enzyme binding and ion transport. In addition, two pathways related to bacterial adhesion were enriched, namely gap junction and adherens junction (Fig. 3b).
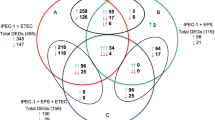
Notably, circRNAs produced by the parental genes involved in the adherens junction pathway were highly expressed in the adhesive group, indicating that these circRNAs may play an important role in regulating the biogenesis of E. coli diarrhea (Fig. 3c). Then we performed an overlap analysis of DE circRNA host genes and DE genes between the adhesive group and non-adhesive group, and obtained eight overlap** genes (Fig. 3a), among which sorbin and SH3 domain-containing protein 1 (SORBS1) attracted our attention due to it was involved in the adherens junction pathway.
Construction of a ceRNA network
It was widely accepted that circRNA can act as a miRNA sponge to directly bind to miRNA, and therefore affect the expression levels of the miRNA target genes [22]. Thus, the present study predicted the targeted miRNAs of the DE circRNAs using the miRanda software, so as to explore the potential molecular sponge function of circRNA. The analysis predicted DE circRNAs binding to 451 porcine mature miRNAs and 6485 circRNA − miRNA interactions (Additional file 1: Table S5). Among them, circ-SORBS1 tends to have a lot of miRNA binding sites, indicating its potential molecular sponge function. Figure 4a and b show part of circ-SORBS1’s miRNA binding sites (Fig. 4a, b). Subsequently, 10 miRNAs were selected to construct the circRNA-miRNA interaction network (Fig. 4c), among which 5 were targeted miRNAs of the key candidate circRNA found in this study, and the other 5 selected miRNAs were found to be related to bacterial diarrhea in previous studies [RNA isolation and quality assessment The total RNA was extracted from small intestine tissues of eight (8) selected piglets by Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction. The kaiaoK5500®Spectrophotometer (Kaiao, Bei**g, China) was adopted to monitor RNA purity. The concentrations of isolated RNA were determined using the Nano Drop spectrophotometer. RNA integrity and concentration was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The quality of all the RNA samples were good enough (OD260/280 > 1.90, RNA integrity number > 8.7) to do the sequencing. Then 20 µL of the isolated total RNA from each sample were sent to company (Annoroad Gene Technology Corporation -Bei**g) for sequencing. A total amount of 2 μg RNA per sample was used as an input material for the RNA library preparations. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (#E7530L, NEB, USA) following the manufacturer's recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. More details about library preparation have been described in our previously published paper [61]. The libraries were sequenced on an Illumina platform (HiSeq Xten) and 150 bp paired-end reads were generated. Raw reads in fastq format were firstly processed through quality control. Briefly, reads containing adapter, polyN and low-quality bases at high proportion were removed to obtain clean reads. The above steps have already been done by sequencing company. All the downstream analyses were based on the clean reads with high quality. Reference genome and annotation files were downloaded from genome website (version: Sscrofa11.1; GCA_000003025.6). The circRNAs were detected by two software of CIRI2 [62] and find_circ [63]. The intersecting results of the two tools were considered as candidate circRNAs and used for subsequent analysis. In this study, the spliced reads per billion map** (SRPBM) method was used to estimate the circRNA expression level. The calculation formula was as follows: Candidate circRNAs were annotated using circAnno software [64]. To identify reverse complementary matches (RCMs), we aligned two introns’ sequences flanking the same exonic circRNA using Basic Local Alignment Search Tool (BLAST) with ‘blastn’ task, ‘-word_size 7’ and ‘-evalue 20’. Briefly, the upstream and downstream introns of an exonic circRNA were input as query sequences and subjected sequences, respectively. For each intron pair, several alignments were obtained, and the alignment with lowest e-value that passed the threshold was regarded as reverse complementary match (RCM). Then we downloaded the repetitive sequences in pig from UCSC Table Browser (http://genome.ucsc.edu/cgi-bin/hgTables) and compared it with RCM genomic coordinates to discover significant overlaps of RCMs and repeat elements. Differential expression (DE) analysis of adhesive and non-adhesive groups was performed using the DESeq2 R package (1.34.0) based on the negative binomial distribution [65]. Candidate circRNAs subject to criteria of p value < 0.05 and |log2(FoldChange)|> 1 were assigned as DE circRNAs between non-adhesive group and adhesive group. Gene ontology (GO) enrichment and KEGG [66] pathway analysis for parental genes of DE circRNAs were conducted by the KOBAS software [67]. GO terms and KEGG pathway with p < 0.05 were considered to be significantly enriched. Combining our previous mRNA data (PRJNA562774, more details can be found in previously published paper [61].) with predicted DE circRNA targeted miRNA data, we constructed a regulatory network of circRNA-miRNA-mRNA to reveal the potential association of ETEC-F4ac adhesion in small intestinal epithelial cells of piglets. All porcine mature miRNAs sequences published in miRbase [68] (http://www.mirbase.org/) were downloaded for further analysis. In details, miRNA binding sites of all DE circRNAs were predicted using miRanda software with free energies of ≤ -20.0 kcal/mol and paring score ≥ 150. Subsequently, target genes of miRNA were predicted using TargetScan (https://www.targetscan.org/vert_80/) and miRnet [69] (https://www.mirnet.ca/) databases. The intersection results of the DE genes that obtained in our mRNA data and predicted target genes were taken as the candidate target genes. Potential circRNA-miRNA-mRNA interactions were established and visualized using Cytoscape 3.9.1 software [70] (http://cytoscape.org/). Gene ontology (GO) enrichment and KEGG pathway analysis for candidate target genes were conducted by the KOBAS software. GO terms and KEGG pathway with p < 0.05 were considered to be significantly enriched.Library preparation for RNA sequencing
Quality control for raw reads and circRNA identification
Differential expression and functional enrichment analysis of circRNA
CircRNA-miRNA-mRNA network analysis
Availability of data and materials
Raw sequencing data for the eight samples analyzed in this study have been uploaded to the Sequence Read Archive (SRA) in National Center for Biotechnology Information (NCBI), and are available under accession number: PRJNA562774 or (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA562774). Data generated during analysis are included in the manuscript as supplementary files.
Abbreviations
- ETEC:
-
Enterotoxigenic Escherichia coli
- F4ab:
-
Fimbriae 4 ab antigen variant
- F4ac:
-
Fimbriae 4 ac antigen variant
- F4ad:
-
Fimbriae 4 ad antigen variant
- ITGB5 :
-
Integrin beta-5
- RNA-seq:
-
RNA-sequencing
- DE:
-
Differentially expressed
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- SORBS1 :
-
Sorbin and SH3 domain-containing protein 1
- E.coli :
-
Escherichia coli
- circRNA:
-
Circular RNA
- miRNA:
-
MicroRNA
- ceRNA:
-
Competitive endogenous RNA
- PEDV:
-
Porcine endemic diarrhea virus
- IPEC-J2:
-
Intestinal Porcine Epithelial Cell line-J2
- SRPBM:
-
Spliced reads per billion map**
- RCMs:
-
Reverse complementary matches
- BLAST:
-
Basic Local Alignment Search Tool
- SINE:
-
Short interspersed element
- LINE:
-
Long interspersed element
- IEC:
-
Intestinal epithelial cells
- DEC:
-
Differentially expressed circRNA
- DEG:
-
Differentially expressed gene
- MYH11 :
-
Myosin heavy chain 11
- MYLK :
-
Myosin light chain kinase
- ITGB1 :
-
Integrin beta-1
- FN1 :
-
Fibronectin
- mRNA:
-
Messenger RNA
References
Ren J, Yan X, Ai H, Zhang Z, Huang X, Ouyang J, Yang M, Yang H, Han P, Zeng W, et al. Susceptibility towards enterotoxigenic Escherichia coli F4ac diarrhea is governed by the MUC13 gene in pigs. PLoS ONE. 2012;7(9):e44573.
Guinée P, Jansen W. Behavior of Escherichia coli K antigens K88ab, K88ac, and K88ad in immunoelectrophoresis, double diffusion, and hemagglutination. Infect Immun. 1979;23(3):700–5.
Geenen P, Van der Meulen J, Bouma A, De Jong M. Estimating transmission parameters of F4+ E. coli for F4-receptor-positive and-negative piglets: one-to-one transmission experiment. Epidemiol Infect. 2004;132(6):1039–48.
Fu W-X, Liu Y, Lu X, Niu X-Y, Ding X-D, Liu J-F, Zhang Q. A genome-wide association study identifies two novel promising candidate genes affecting Escherichia coli F4ab/F4ac susceptibility in swine. PLoS ONE. 2012;7(3):e32127.
Wang W, Liu Y, Tang H, Yu Y, Zhang Q. ITGB5 plays a key role in Escherichia coli F4ac-induced diarrhea in piglets. Front Immunol. 2019;10:2834.
Zhang P, Chao Z, Zhang R, Ding R, Wang Y, Wu W, Han Q, Li C, Xu H, Wang L. Circular RNA regulation of myogenesis. Cells. 2019;8(8):885.
Chen L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–8.
Robic A, Cerutti C, Kuhn C, Faraut T. Comparative analysis of the circular transcriptome in muscle, liver, and testis in three livestock species. Front Genet. 2021;12:665153.
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
Li M, Zhang N, Zhang W, Hei W, Cai C, Yang Y, Lu C, Gao P, Guo X, Cao G, et al. Comprehensive analysis of differentially expressed circRNAs and ceRNA regulatory network in porcine skeletal muscle. BMC Genomics. 2021;22(1):320.
Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7(30):47186.
Li B, He Y, Wu W, Tan X, Wang Z, Irwin DM, Wang Z, Zhang S. Circular RNA profiling identifies novel circPPARA that promotes intramuscular fat deposition in pigs. Agric Food Chem. 2022;70(13):4123–37.
Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588–600.
Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237–45.
Li T-R, Jia Y-J, Wang Q, Shao X-Q, Lv R-J. Circular RNA: a new star in neurological diseases. Int J Neurosci. 2017;127(8):726–34.
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–28.
Xu S, Zhou L, Ponnusamy M, Zhang L, Dong Y, Zhang Y, Wang Q, Liu J, Wang K. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ. 2018;6:e5503.
Yan Z, Jiang T, Wang P, Huang X, Yang Q, Sun W, Gun S. Circular RNA expression profile of spleen in a Clostridium perfringens type C-induced piglet model of necrotizing enteritis. FEBS Open Bio. 2018;8(10):1722–32.
Chen J, Wang H, ** L, Wang L, Huang X, Chen W, Yan M, Liu G. Profile analysis of circRNAs induced by porcine endemic diarrhea virus infection in porcine intestinal epithelial cells. Virology. 2019;527:169–79.
Liu Y-C, Li J-R, Sun C-H, Andrews E, Chao R-F, Lin F-M, Weng S-L, Hsu S-D, Huang C-C, Cheng C. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–15.
Wu W, Ji P, Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020;21(1):1–14.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8.
Wang W, Wang P, **e K, Luo R, Gao X, Yan Z, Huang X, Yang Q, Gun S. ssc-miR-185 targets cell division cycle 42 and promotes the proliferation of intestinal porcine epithelial cell. Animal Bioscience. 2021;34(5):801.
Sun L, Wu S, Dai C-H, Sun S-Y, Zhu G-Q, Wu S-L, Bao W-B. Insight into the molecular mechanism of miR-192 regulating Escherichia coli resistance in piglets. Biosci Rep. 2018;38(1):BSR20171160.
Luo R, Yan Z, Yang Q, Huang X, Gao X, Wang P, Wang W, **e K, Gun S. Inhibition of ssc-microRNA-140-5p ameliorates the Clostridium perfringens beta2 toxin-induced inflammatory response in IPEC-J2 cells via the ERK1/2 and JNK pathways by targeting VEGFA. Mol Immunol. 2020;127:12–20.
Wang W, Yang Q, Huang X, Luo R, **e K, Gao X, Yan Z, Wang P, Zhang J, Yang J. Effects of miR-204 on apoptosis and inflammatory response of Clostridium perfringens beta2 toxin induced IPEC-J2 cells via targeting BCL2L2. Microb Pathog. 2021;156:104906.
Zheng H, Xu L, Liu Y, Li C, Zhang L, Wang T, Zhao D, Xu X, Zhang Y. MicroRNA-221-5p inhibits porcine epidemic diarrhea virus replication by targeting genomic viral RNA and activating the NF-κB pathway. Int J Mol Sci. 2018;19(11):3381.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
Aijaz S, Balda MS, Matter K. Tight junctions: molecular architecture and function. Int Rev Cytol. 2006;248:261–98.
Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809.
McCole DF, Barrett KE. Varied role of the gut epithelium in mucosal homeostasis. Curr Opin Gastroenterol. 2007;23(6):647–54.
Awad WA, Hess C, Hess M. Enteric pathogens and their toxin-induced disruption of the intestinal barrier through alteration of tight junctions in chickens. Toxins. 2017;9(2):60.
Fasano A, Nataro JP. Intestinal epithelial tight junctions as targets for enteric bacteria-derived toxins. Adv Drug Deliv Rev. 2004;56(6):795–807.
Dytoc M, Fedorko L, Sherman PM. Signal transduction in human epithelial cells infected with attaching and effacing Escherichia coli in vitro. Gastroenterology. 1994;106(5):1150–61.
Xue Y, Du M, Sheng H, Hovde CJ, Zhu M-J. Escherichia coli O157: H7 suppresses host autophagy and promotes epithelial adhesion via Tir-mediated and cAMP-independent activation of protein kinase A. Cell death discovery. 2017;3(1):1–11.
Yuhan R, Koutsouris A, Savkovic SD, Hecht G. Enteropathogenic Escherichia coli-induced myosin light chain phosphorylation alters intestinal epithelial permeability. Gastroenterology. 1997;113(6):1873–82.
Dubreuil JD. Enterotoxigenic Escherichia coli targeting intestinal epithelial tight junctions: an effective way to alter the barrier integrity. Microb Pathog. 2017;113:129–34.
Guttman JA, Lin AEJ, Li Y, Bechberger J, Naus CC, Vogl AW, Finlay BB. Gap junction hemichannels contribute to the generation of diarrhoea during infectious enteric disease. Gut. 2010;59(2):218–26.
Oda H, Takeichi M. Structural and functional diversity of cadherin at the adherens junction. J Cell Biol. 2011;193(7):1137–46.
Germain M, Pezzolesi MG, Sandholm N, McKnight AJ, Susztak K, Lajer M, Forsblom C, Marre M, Parving H-H, Rossing P. SORBS1 gene, a new candidate for diabetic nephropathy: results from a multi-stage genome-wide association study in patients with type 1 diabetes. Diabetologia. 2015;58(3):543–8.
Lin W-H, Chiu KC, Chang H-M, Lee K-C, Tai T-Y, Chuang L-M. Molecular scanning of the human sorbin and SH3-domain-containing-1 (SORBS1) gene: positive association of the T228A polymorphism with obesity and type 2 diabetes. Hum Mol Genet. 2001;10(17):1753–60.
Cho WC, Jang JE, Kim KH, Yoo BC, Ku JL. SORBS1 serves a metastatic role via suppression of AHNAK in colorectal cancer cell lines. Int J Oncol. 2020;56(5):1140–51.
** X, Qiu X, Huang Y, Zhang H, Chen K. miR-223-3p carried by cancer-associated fibroblast microvesicles targets SORBS1 to modulate the progression of gastric cancer. Cancer Cell Int. 2022;22(1):1–15.
Li X, Wubbolts RW, Bleumink-Pluym NM, van Putten JP, Strijbis K. The transmembrane mucin MUC1 facilitates β1-integrin-mediated bacterial invasion. MBio. 2021;12(2):e03491-e3420.
Isberg RR, Leong JM. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell. 1990;60(5):861–71.
Ljungh A, Emödy L, Steinruck H, Sullivan P, West B, Zetterberg E, Wadström T. Fibronectin, vitronectin, and collagen binding to Escherichia coli of intestinal and extraintestinal origin. Zentralbl Bakteriol. 1990;274(1):126–34.
Wang P, Huang X, Yan Z, Yang Q, Sun W, Gao X, Luo R, Gun S. Analyses of miRNA in the ileum of diarrheic piglets caused by Clostridium perfringens type C. Microb Pathog. 2019;136:103699.
Tan B, Li Y, Zhao Q, Fan L, Wang D. ZNF139 increases multidrug resistance in gastric cancer cells by inhibiting miR-185. Biosci Rep. 2018;38(5):BSR20181023.
Yuan M, Zhang X, Zhang J, Wang K, Zhang Y, Shang W, Zhang Y, Cui J, Shi X, Na H. DC-SIGN–LEF1/TCF1–miR-185 feedback loop promotes colorectal cancer invasion and metastasis. Cell Death Differ. 2020;27(1):379–95.
Wang Y-P, Huang Y, Hou T, Lu M. LncRNA XIST acts as a ceRNA sponging miR-185-5p to modulate pancreatic cancer cell proliferation via targeting CCND2. Transl Cancer Res. 2020;9(3):1427.
Liu Y, Chen S-H, ** X, Li Y-M. Analysis of differentially expressed genes and microRNAs in alcoholic liver disease. Int J Mol Med. 2013;31(3):547–54.
Maruyama S, Furuya S, Shiraishi K, Shimizu H, Akaike H, Hosomura N, Kawaguchi Y, Amemiya H, Kawaida H, Sudo M. miR-122-5p as a novel biomarker for alpha-fetoprotein-producing gastric cancer. World J Gastrointest Oncol. 2018;10(10):344.
Meng L, Chen Z, Jiang Z, Huang T, Hu J, Luo P, Zhang H, Huang M, Huang L, Chen Y. MiR-122-5p suppresses the proliferation, migration, and invasion of gastric cancer cells by targeting LYN. Acta Biochim Biophys Sin. 2020;52(1):49–57.
Xu Z, Liu G, Zhang M, Zhang Z, Jia Y, Peng L, Zhu Y, Hu J, Huang R, Sun X. miR-122-5p inhibits the proliferation, invasion and growth of bile duct carcinoma cells by targeting ALDOA. Cell Physiol Biochem. 2018;48(6):2596–606.
Wei Q, Tu Y, Zuo L, Zhao J, Chang Z, Zou Y, Qiu J. MiR-345-3p attenuates apoptosis and inflammation caused by oxidized low-density lipoprotein by targeting TRAF6 via TAK1/p38/NF-kB signaling in endothelial cells. Life Sci. 2020;241:117142.
Li Y, Wu X, Li J, Du L, Wang X, Cao J, Li H, Huo Z, Li G, Pan D. Circ_0004354 might compete with circ_0040039 to induce NPCs death and inflammatory response by targeting miR-345–3p-FAF1/TP73 axis in intervertebral disc degeneration. Oxid Med Cell Longev. 2022;2022:2776440.
Loos M, Geens M, Schauvliege S, Gasthuys F, van der Meulen J, Dubreuil JD, Goddeeris BM, Niewold T, Cox E. Role of heat-stable enterotoxins in the induction of early immune responses in piglets after infection with enterotoxigenic Escherichia coli. PLoS ONE. 2012;7(7):e41041.
Augustino SMA, Xu Q, Liu X, Mi S, Shi L, Liu Y, Wen H, Wang D, Liu L, Zhang Q, et al. Integrated analysis of lncRNAs and mRNAs reveals key trans-target genes associated with ETEC-F4ac adhesion phenotype in porcine small intestine epithelial cells. BMC Genomics. 2020;21(1):780.
Li Y, Qiu X, Li H, Zhang Q. Adhesive patterns of Escherichia coli F4 in piglets of three breeds. J Genet Genomics. 2007;34(7):591–9.
Xu Q-l. Serafino M, Agustino A, Si-yuan M, Ya-chun W, **-xia H: Differential expression analysis of ITGB5 and MUC13 genes in ETEC resistance and susceptible large white piglets. Chin J Anim Sci. 2019;55(2):39–43.
Augustino S, Xu Q, Liu X, Liu L, Zhang Q, Yu Y. Transcriptomic study of porcine small intestine epithelial cells reveals important genes and pathways associated with susceptibility to Escherichia coli F4ac diarrhea. Front Genet. 2020;11:68.
Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19(5):803–10.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–7.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
**e C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(suppl_2):W316–22.
Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–62.
Chang L, Zhou G, Soufan O, **a J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(1):244–51.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Acknowledgements
We thank all co-authors for their contributions and Yu Zhang for assistance in the analysis and processing of RNA-seq data.
Funding
This research was financially funded by the National Natural Science Foundation (NSFC31572361). The program for Chang Jiang Scholar and Innovation Research Team in University (IRT-15R62). The National Key Research and Development Project (2019YFE0106800). Funding body had no contribution in the conception and the design of the study, sample collection, analysis and interpretation of data and in the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
QYZ analyzed the data and wrote the manuscript. SMA participated in results discussion and analyzed RNA-seq data. QX assisted in performing some of the experiments and gathered literature. CW participated in the result interpretation and paper revision. YY and QZ conceived and designed the study and revised the manuscript. All the authors contributed to manuscript revisions, read and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The animal study and all animal procedures were approval by the Animal Welfare Committee of China Agricultural University. The experiments were conducted following the protocol approved by the Animal Welfare Committee of China Agricultural University (Permit Number: DK996). All methods were performed in accordance with the relevant guidelines and regulations. This study was carried out in compliance with the ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
All authors declared no competing interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Summary of circRNAs identified in eight samples. Table S2. The counts of the back-spliced junction read of 8 samples were normalized as the spliced reads per billion map** (SRPBM). Table S3. Differentially expressed circRNAs (DEC) between adhesive and non-adhesive group. Table S4. Significantly enriched GO terms of DE circRNA host genes between adhesive and non-adhesive group. Table S5. Predicted DE circRNA's miRNA binding sites. Table S6. Significantly enriched GO terms and KEGG pathways of DE genes between adhesive and non-adhesive group.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhao, Q., Xu, Q., Serafino, M. et al. Comprehensive analysis of circular RNAs in porcine small intestine epithelial cells associated with susceptibility to Escherichia coli F4ac diarrhea. BMC Genomics 24, 211 (2023). https://doi.org/10.1186/s12864-022-08994-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08994-8