Abstract
Background
RNA velocity is a novel and powerful concept which enables the inference of dynamical cell state changes from seemingly static single-cell RNA sequencing (scRNA-seq) data. However, accurate estimation of RNA velocity is still a challenging problem, and the underlying kinetic mechanisms of transcriptional and splicing regulations are not fully clear. Moreover, scRNA-seq data tend to be sparse compared with possible cell states, and a given dataset of estimated RNA velocities needs imputation for some cell states not yet covered.
Results
We formulate RNA velocity prediction as a supervised learning problem of classification for the first time, where a cell state space is divided into equal-sized segments by directions as classes, and the estimated RNA velocity vectors are considered as ground truth. We propose Velo-Predictor, an ensemble learning pipeline for predicting RNA velocities from scRNA-seq data. We test different models on two real datasets, Velo-Predictor exhibits good performance, especially when XGBoost was used as the base predictor. Parameter analysis and visualization also show that the method is robust and able to make biologically meaningful predictions.
Conclusion
The accurate result shows that Velo-Predictor can effectively simplify the procedure by learning a predictive model from gene expression data, which could help to construct a continous landscape and give biologists an intuitive picture about the trend of cellular dynamics.
Similar content being viewed by others
Background
Recent advances in high-throughput RNA sequencing technologies [1] have enabled analysis of transcription at single-cell level [2], which has provided immense opportunities to unravel the underlying mechanisms of gene expression regulation. However, in many cases, dynamical information of cell state transition is limited. When the sequencing is completed, the expression data provide only a snapshot of a cell [3]. Currently, trajectory inference (including pseudotime analysis) is a primary task to identify cells in various states of differentiation [4]. In general, trajectory inference methods need to construct graphs. There are various approaches to trajectory reconstrcution, e.g. SCUBA [5] is based on bifurcation analysis, SCENT [6] and scEpath [7] use a measurement of entropy of cell states. HopLand [8] and Topslam [9] project cells to a landscape with optimized parameters.
A major limitation of most trajectory inference methods [10] is they do not connect data to underlying molecular kinetics. La Manno et al. found that spliced and unspliced mRNAs can be distinguished in standard single-cell RNA-seq protocols [11], and the timescale of differentiation during development is comparable to the typical half-life of an mRNA. Hence, we can use the abundances of mRNAs to estimate splicing rate and degradation rate. They proposed a simple kinetic framework for estimating changes in mRNA levels of individual cells. This framework is based on the central dogma of molecular biology. Gorini and Maas proposed a first-order differential equation to model this biological process [12], to which Zeisel et al. added intermediate steps [13].
The original steady-state model for RNA velocity proposed by La Manno et al. assumes that transcriptional phases endure long enough to reach a steady state equilibrium, and the equilibrium mRNA levels can be approximated with a linear regression by simplification with a common splicing rate. Recently, to relax this assumption, Volker Bergen et al. proposed an algorithm called “scVelo” [14], which includes a stochastic model and a dynamical model in addition to the steady-state model. The stochastic model treats the transcription, splicing and degradation as probabilistic events, which means steady-state levels are approximated not only from mRNA levels, but also from intrinsic expression variability. The dynamical model considers non-stationary populations and different splicing rates across genes, and the dynamics is solved in a maximum likelihood framework using the expectation maximization (EM) algorithm. The dynamical model is slower but can provide more consistent velocity estimation and better identification of transcriptional states.
The concept of RNA velocity and its associated algorithms and models have become very popular in single-cell biology. However, this technique needs the support of RNA sequencing protocols. Moreover, to get splicing information we need to run a complex preprocessing pipeline which involves the issues of file format and is time consuming. More importantly, the data of estimated RNA velocities are still sparse compared with the size of the uncovered cell state space. Here we propose an ensemble learning pipeline for the prediction of RNA velocities, which can skip the complex procedures for splicing analysis, etc. When we have a new data sample from the same biological context, we can predict the direction of RNA velocity from a state unkown in the traning data. This is similar to the pedestrian prediction [15] in a driverless transportation system, or the prediction of next movements of basketball players on the court [16]. It is possible to further combine all the transient movements into long trajectories of cells. Inspired by the concept of Waddington’s epigenetic landscape, which is a classical metaphor for cell differentiation, we can treat cells as balls rolling down through a potential surface. Based on the predicted RNA velocities and cell trajectories, we can reconstruct the landscape, as an intuitive platform for single-cell data visualization.
Methods
Velocity estimation
Velocyto CLI or loompy/kallisto was used to obtain spliced/unspliced reads annotations. We filter the genes with counts number (both spliced and unspliced) smaller than the threshold, keep the top high variability genes. Then normalize in cell level and did logarithm transform. On Euclidean distances PCA space of counts matrix, a nearest neighbor graph was computed, first and second moments were obtained for each cell. According to the basic reaction kinetics:
where S(t) represents mature mRNA abundance over time, U(t) represents pre-mRNA abundance over time, \(\alpha\) is the rate of transcription, \(\beta\) is the rate of splicing, and \(\gamma\) is the rate of degradation. k and t are cell-specific latent variables, where k represents discrete transcriptional state, and t represents latent time.
RNA velocity is termed as the time derivative of mature spliced mRNA \(v(t) = \frac{dS(t)}{dt}\). Three approaches are provided in scVelo to do velocity estimation: steady state model, stochastic model and dynamical model. The basic difference between them is that the assumptions about the parameters are different. The data preprocessing steps are shown in Algorithm 1. For the sake of completeness and readers’ convenience, we have rephrased their description of methods for RNA velocity estimation into the pseudocode. After velocity estimation we can get a multi-dimensional RNA velocity vector V for each transcriptional state of a single cell. Combining this information we can further inference cell future state of an individual cell. The movements can be UMAP projected into a lower dimensional embedding D to visualize.

Problem statement
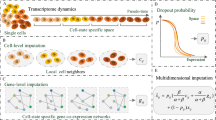
Our goal is to predict the RNA velocity vector of each cell based on its gene expression data. As illustrated in a 2D space, we formulate it as a classification problem through an equal division of a 2D circle into d equal-sized segments, as shown in Fig. 1. If the predicted and the original target directions fall in the same segment, we count it as a true positive, etc. A slightly more realistic formulation of the problem could be the regression of angles of the RNA velocities from a fixed direction. But we will leave that as a future work.
Input preprocessing
Figure 2 shows the whole pipeline of our work. We start our supervised learning task from the gene expression matrix. Single-cell genomic data may be sparse and suffer from technical noise and bias. Therefore, to improve the behavior we need to do a de-noising step, or called feature engineering step. There are several ways to do such as scVI [17], scVAE [18] and DCA [19]. They basically use auto-encoder to find the hidden layer with minimum reconstruction error. After comparison we do feature selection based on gene ranking by ScVelo. ScVelo ranking is based on cluster-specific t-test to find genes with significantly higher/lower differential velocity, and we select the top k genes from each cluster as the model features. We use the lower-dimensional embedding \(D\in \mathbf {R}^{2n}\) obtained from the velocity estimation step as our ground truth.
After the division, the distribution of labels is not balanced. Therefore, we provide several ways to rescue: over-sampling, down-sampling and combine-sampling. We first test different sampling ways on different base models. For oversampling, we test adaptive synthetic (ADASYN) [20], synthetic minority oversampling technique (SMOTE) [21] and some variants of SMOTE, such as border line smote (BLS) [22] and svm-smote which uses support vector machine (SVM) [23]. For down-sampling, cluster centroid (CC) [24], random under sampler (RUS), NearMiss [25], repeated edited nearest neighbours (RENN) [26], neighbourhood cleaning rule (NCR) [27] and one side selection (OSS) [28] are used. Then, we further test combine-sampling methods of SMOTETomek [29] and SMOTEENN [30].
Model training
We divide the sample data into a training set and test set. The training set is for model training and test set is for model evaluation. For training, the parameters are saved and can be directly used for prediction in testing part. We adapt a stacking structure model. Figure 3 and Algorithm 2 show the detail. We use mlxtend [31] and Scikit-learn [32] packages for implementation. We choose random forest (RF), GBDT, extra tree classifier (ET), adaboost (ADA) and XGBoost model to be the first layer, and the second layer is a simple Logistic regression classifier. To avoid over-fitting, we use cross validation concept to divide the training set to K subsets, where \(K-1\) subsets are used to fit the first layer of classifiers. Then in each round, the unused subset will be predicted by the fitted classifier, and all the resulting predictions are stacked to feed into the second layer.

Results
Data sets
We train and test the models on two single-cell RNA-seq datasets. One is the Mouse hippocampal dentate gyrus neurogenesis (DGN) dataset [33] available from NCBI Gene Expression Omnibus (GEO) under accession ID GSE95753. It consists of RNA-seq data of 13,913 genes and 2930 cells from multiple lineages. The other dataset is Pancreatic endocrinogenesis (PE) [34] also available from NCBI GEO under accession ID GSE132188, which comprises the transcriptional levels of 27,998 genes of 3,696 pancreatic epithelial and Ngn3-Venus fusion cells sampled from mouse embryonic day 15.5. The number of cells and the numbers of genes (with different values of k, the number of top genes selected from each cluster as features) are shown in Table 1.
To test the generalization ability of our models, we randomly divide the cell samples into two disjoint sets with ratio of 7:3, 7 for training and 3 for testing. Figure 4a shows the proportion of labels in the DG datasets.
Class imbalance issue
To illustrate how to address the class imbalance issue, we take the DG dataset as an example. Figure 4a shows the label proportion of the DG dataset. Figure 4b–d shows the ROC curve and corresponding AUC score of different sampling strategies. The AUC score is not enough for imbalanced data, thus we also consider the precision on each of the four classes and the balanced score as metrics. The metrics can be calculated according to the following equations:
Table 2 shows the most representative performance of each methods. We can see the down-sampling methods perform poorly because of loss of information. The over-sampling methods are better but may introduce some biases. The best way is to combine them. Therefore we choose SMOTETomek as our final choice for the DG dataset.
Functional analysis
After parameters fine tuning through grid search, Fig. 5 visualizes the performances of base models and stacking model. Figure 6a shows the loss curve on first fold, the behaviors of the other folds are similar. XGBoost model can also provide the log loss curve and the most important genes learning from the data (Fig. 6b). The impact of hyper parameters k the number of top genes and d the number classes is shown in Fig. 7b. Parameter k controls the feature selection part. The curve rises first and after k reaches 20, it starts to oscillate. In the previous experiment we set k to 3, although we can increase k to get better performance. Parameter d controls the granularity of prediction, and the result shows that the number of divisions is 8. When we continue to increase d, the task becomes more difficult so that the score will decay. Figure 7a shows the best performance of stacking model on DG dataset with \(d = 8\), \(k = 20\). Table 3 shows the comparison of base models and final stacking model. Stacking model’s performance is even slightly worse than XGBoost. We think that is probably due to the following reasons. First, the dataset is too small because stacking is not so strong when the data set is not big enough. Secondly, we can increase the model diversity of the first layer, and add more models to improve the performance. Thirdly, we did not use cross validation technique in the random forest model. In conclusion, we think the score difference is small and users can choose different provided models according to their size of data set.
Confusion matrix of different models. Here shows the case when the number of classes d equals 4 on the DG dataset. a The performance of the XGBoost model, where each square corresponds to a predicted label and a true label for direction of RNA velocity, and the value in each square is the number of cells meeting the condition (color becomes darker as the number increases). We can see that the diagnals have darker colors which means that some samples are correctly classified. b The performance of the Random Forest model. c The performance of the Stacking model
Visualization
We use the UMAP toolkit of scVelo [14] to map cells and velocity vectors into two dimensional space, and we assume the incoming new data has the same distribution with our training data. We projected the new data in the same way as previous embedding, and give them a small red arrow which indicates the prediction of our velocity direction information. Each point in the figure represents a cell, the arrow is the velocity information of cells. It gives us an intuitive instruction of which way where a specific cell goes to. In Biology, it will tell us the differential path of a cell, we can see it performs well. Figure 8 shows the result on dataset PE, different colors indicate different clusters, above figure is the ground truth. In below figure, red arrow is our prediction outcome. Comparing with the same location in ground truth figure, we can see that the outcome is consistent with the ground truth. For detail, the orange dots represent pre-endocrine cells, and the perple dots represent epsilon cells. Through the zoom-in window, the comparision shows clearly that cell movements on 2D space are well captured.
The impact of hyperparameters on DG dataset. a Balance score curve with different hyperparameters. The upper one shows the balance score increases with the increasing top gene number k. After k reaches 20, the balance score begins to oscillate which means 20 is enough for a good performance. The bottom one shows the performance drops when the number of classes exceeds 8. b Confusion matrix of the Stacking model when the number of genes k equals 20 and the number of classes d equals to 8. Each square corresponds to predicted label and a true label, and the value in each square is the number of cells satisfying the condition (color is darker as the number increases). The numbers along the diagnal show the performance of the Stacking model on DG dataset
Schematic diagram of movement prediction on PE dataset. The leftside figure shows the velocity distribution of PE dataset, where different colors indicate different clusters. In the zoom-in figures on the right side, the upper one shows cells and velocity directions with original labels (training set), and in the bottom figure, the red arrows without halos represent cells and velocity directions with predicted labels (test set)
Discussion
One limitation of Velo-Predictor is that its performance may depend on data. Although we have used the ensemble learning framework to balance the results from different baseline models for different sample-feature ratios, it is still an empirical approach. When the data distribution is not so complete and balanced, the prediction may be less accurate. The lowest input data size depends on the properties of data (e.g. in terms of samples and features) and biological scenarios (e.g. types of data). We have tested the case (\(k = 5\), \(d = 4\)) on the DG dataset by adjusting the testing data size, and when it exceeds 40%, the number of errors will increase to hundreds. In general, more complete data that cover the dynamical processes in the biological scenarios under study would be much preferred. Besides, the interpretability of the model is still a challenge.
Our work provides a prediction-based approach for study of cell differentiation mechanisms. Such predictions can also help impute the state space not yet covered by the scRNA-seq data, and the interpolation can help construct a continuous landscape surface. In the future, we can use single-cell multi-omic data to learn the bifucation point of cell differentiation more precisely. With the imputed direction information we can further do trajectory inference. Combined with an energy function such as that in the Hopfield network model used in our previous work [8], the predicted RNA velocities can be used to model the Waddington’s epigenetic landscape. Therefore, the prediction of RNA velocity can give biologists an intuitive picture about the trend of cellular dynamics, which is informative for their research.
Conclusion
In this paper, we described Velo-Predictor, an ensemble learning pipeline for RNA velocity prediction. While RNA velocity estimation is not straightforward, our pipeline can simplify the procedure by learning a predictive model from gene expression data. The results showed that our pipeline can predict the directions of cell state transitions accurately.
Availability of data and materials
The single cell RNAseq data are publicly available in Gene Expression Omnibus, accession numbers GSE95753 and GSE132188. The code is available at https://github.com/clay001/Velo-Predictor.
Abbreviations
- scRNA-seq:
-
Single-cell RNA sequencing
- EM:
-
Expectation maximization
- ADASYN:
-
Adaptive synthetic
- SMOTE:
-
Synthetic minority over sampling technique
- BLS:
-
Border line smote
- SVM:
-
Support vector machine variant of smote
- CC:
-
Cluster centroid
- RUS:
-
Random under sampler
- RENN:
-
Repeated edited nearest neighbours
- NCR:
-
Neighbourhood cleaning rule
- OSS:
-
One side selection
- RF:
-
Random forest
- GBDT:
-
Gradient boosting decision tree
- ET:
-
Extra tree classifier
- ADA:
-
Adaboost
- DGN:
-
Dentate gyrus neurogenesis
- GEO:
-
Gene Expression Omnibus
- PE:
-
Pancreatic endocrinogenesis
References
Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8(1):1–12.
Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13(4):599–604.
Weinreb C, Wolock S, Tusi BK, Socolovsky M, Klein AM. Fundamental limits on dynamic inference from single-cell snapshots. Proc Natl Acad Sci. 2018;115(10):2467–76.
Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37(5):547–54.
Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan G-C. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci. 2014;111(52):5643–50.
Teschendorff AE, Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome. Nat Commun. 2017;8(1):1–15.
** S, MacLean AL, Peng T, Nie Q. scEpath: energy landscape-based inference of transition probabilities and cellular trajectories from single-cell transcriptomic data. Bioinformatics. 2018;34(12):2077–86.
Guo J, Zheng J. HopLand: single-cell pseudotime recovery using continuous hopfield network-based modeling of Waddington’s epigenetic landscape. Bioinformatics. 2017;33(14):102–9.
Zwiessele M, Lawrence ND. Topslam: Waddington landscape recovery for single cell experiments. BioRxiv. 2016;057778.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381.
La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. RNA velocity of single cells. Nature. 2018;560(7719):494–8.
Gorini L, Maas WK. The potential for the formation of a biosynthetic enzyme in Escherichia coli. Biochim Biophys Acta. 1957;25(1):208.
Zeisel A, Köstler WJ, Molotski N, Tsai JM, Krauthgamer R, Jacob-Hirsch J, Rechavi G, Soen Y, Jung S, Yarden Y, et al. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Mol Syst Biol. 2011;7(1):529.
Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–14.
Alahi A, Goel K, Ramanathan V, Robicquet A, Fei-Fei L, Savarese S. Social lstm: human trajectory prediction in crowded spaces. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. p. 961–71.
Felsen P, Lucey P, Ganguly S. Where will they go? Predicting fine-grained adversarial multi-agent motion using conditional variational autoencoders. In: Proceedings of the European conference on computer vision (ECCV); 2018. p. 732–47.
Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15(12):1053–8.
Grønbech CH, Vording MF, Timshel PN, Sønderby CK, Pers TH, Winther O. scVAE: variational auto-encoders for single-cell gene expression datas. BioRxiv; 2018:318295.
Eraslan G, Simon LM, Mircea M, Mueller NS, Theis FJ. Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun. 2019;10(1):1–14.
He H, Bai Y, Garcia EA, Li S. ADASYN: adaptive synthetic sampling approach for imbalanced learning. In: 2008 IEEE international joint conference on neural networks (IEEE world congress on computational intelligence). IEEE; 2008. p. 1322–28.
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–57.
Han H, Wang W-Y, Mao B-H. Borderline-smote: a new over-sampling method in imbalanced data sets learning. In: International conference on intelligent computing. Springer; 2005. p. 878–887.
Nguyen HM, Cooper EW, Kamei K. Borderline over-sampling for imbalanced data classification. Int J Knowl Eng Soft Data Paradigms. 2011;3(1):4–21.
Yuwono M, Su SW, Moulton B, Nguyen H. Fast unsupervised learning method for rapid estimation of cluster centroids. In: 2012 IEEE congress on evolutionary computation. IEEE; 2012. p. 1–8 .
Mani I, Zhang I. kNN approach to unbalanced data distributions: a case study involving information extraction. In: Proceedings of workshop on learning from imbalanced datasets. vol 126; 2003.
Tomek I, et al. An experiment with the edited nearest-nieghbor rule; 1976.
Laurikkala J. Improving identification of difficult small classes by balancing class distribution. In: Conference on artificial intelligence in medicine in Europe. Springer; 2001, p. 63–6.
Kubat M, Matwin S, et al. Addressing the curse of imbalanced training sets: one-sided selection. In: Icml, vol. x97; 1997. p. 179–86. Citeseer
Batista GE, Prati RC, Monard MC. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explor Newsl. 2004;6(1):20–9.
Batista GE, Bazzan AL, Monard MC. Balancing training data for automated annotation of keywords: a case study. In: WOB; 2003. p. 10–18.
Mlxtend Raschka S. providing machine learning and data science utilities and extensions to python’s scientific computing stack. J Open Source Softw. 2018;3(24):638 (https://doi.org/10.21105/joss.00638).
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30.
Hochgerner H, Zeisel A, Lönnerberg P, Linnarsson S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nature Neurosci. 2018;21(2):290–9.
Bastidas-Ponce A, Tritschler S, Dony L, Scheibner K, Tarquis-Medina M, Salinno C, Schirge S, Burtscher I, Böttcher A, Theis FJ, et al. Comprehensive single cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development. 2019;146(12):dev173849.
Acknowledgements
Not applicable.
About this supplement
This article has been published as part of BMC Bioinformatics Volume 22 Supplement 10 2021: Selected articles from the 19th Asia Pacific Bioinformatics Conference (APBC 2021): bioinformatics. The full contents of the supplement are available at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-22-supplement-10.
Funding
This research was supported by a startup grant from the ShanghaiTech University. Publication costs are funded by the same startup grant. The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
XW and JZ conceived the project. XW developed the method and did the experiments. XW and JZ wrote the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, X., Zheng, J. Velo-Predictor: an ensemble learning pipeline for RNA velocity prediction. BMC Bioinformatics 22 (Suppl 10), 419 (2021). https://doi.org/10.1186/s12859-021-04330-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-021-04330-1