Abstract
Background
Quantitative analysis of nanoparticle uptake at the cellular level is critical to nanomedicine procedures. In particular, it is required for a realistic evaluation of their effects. Unfortunately, quantitative measurements of nanoparticle uptake still pose a formidable technical challenge. We present here a method to tackle this problem and analyze the number of metal nanoparticles present in different types of cells. The method relies on high-lateral-resolution (better than 30 nm) transmission x-ray microimages with both absorption contrast and phase contrast -- including two-dimensional (2D) projection images and three-dimensional (3D) tomographic reconstructions that directly show the nanoparticles.
Results
Practical tests were successfully conducted on bare and polyethylene glycol (PEG) coated gold nanoparticles obtained by x-ray irradiation. Using two different cell lines, EMT and HeLa, we obtained the number of nanoparticle clusters uptaken by each cell and the cluster size. Furthermore, the analysis revealed interesting differences between 2D and 3D cultured cells as well as between 2D and 3D data for the same 3D specimen.
Conclusions
We demonstrated the feasibility and effectiveness of our method, proving that it is accurate enough to measure the nanoparticle uptake differences between cells as well as the sizes of the formed nanoparticle clusters. The differences between 2D and 3D cultures and 2D and 3D images stress the importance of the 3D analysis which is made possible by our approach.
Similar content being viewed by others
Background
Quantitative analysis is an important but still largely unexplored issue in the study of nanomedicine procedures, in particular at the cellular and subcellular levels. Many phenomena were discovered by which nanoparticles enhance the cancer cell mortality or facilitate the action of other cell-killing factors [1–4]. However, the potential modulation of these phenomena for procedures such as radiotherapy [5–9] or drug delivery [7, Full size image
The same figure shows that the PEG coating increased (by 30-40%) the nanoparticle damage at very high concentrations. At low concentrations (0.1 mM), the nanoparticles did not significantly affect the cell viability.
To determine if apoptosis was the cause of cell death for highly concentrated PEG-coated AuNPs, we performed flow cytometry with a fluorescence-activated cell sorter (FACS) [41]. As shown in Figure 2, there was no significant increase in the apoptotic cells as the PEG-coated AuNP concentration increased: the profile is similar to that of the control specimen. This indicates that cell death does not occur via apoptosis but via necrosis.
Results of the flow cytometry. The flow cytometry profile of the EMT cell cycle after co-culturing with PEG-coated AuNPs with different colloidal concentrations was performed with a fluorescence-activated cell sorter (FACS). There was no significant increase in the apoptotic cells as the nanoparticle concentration increased (A: Control, B: 0.1 mM, C: 0.5 mM, D: 1.0 mM), indicating that the apoptosis is not likely to cause the observed cell damage in this case.
TEM imaging
TEM was first performed on thin sections, of thickness <100 nm, of all the cell lines with and without exposure to nanoparticle colloid. Figure 3 shows examples of TEM micrographs of the EMT and HeLa cells with AuNPs internalized. After co-culturing with 500 μM naked AuNPs for 48 h, the endocytotic vesicles inside the cytoplasm of cell lines contained clusters of many nanoparticles. However, there are visible differences between the two lines: the vesicle size for EMT cells (A) is substantially larger than for HeLa cells (B).
TEM images of cells with internalized AuNPs. After co-culturing with 500 μM colloidal naked AuNPs for 48 h, the endocytotic vesicles of these cells were found to contain clusters of many AuNPs inside the cytoplasm. Note that the size of the clusters is significantly larger for EMT cells (A and B) than for HeLa cells (C). Bars: 1 μm (A), 200 nm (B) and 5 μm (C).
This is an example of the quantitative information yielded by TEM: the average size of the vesicles in Figure 3A, 3B and 3C are 637 ± 41 nm 530 ± 16 nm and 280 ± 30 nm (n = 5 for EMT cell; n = 4 for HeLa cell). There are, however, limitations in the quantitative data that can be extracted with TEM. The images of Figure 3 are from very thin slices of cells hundreds time thicker, and essentially yield 2D information.
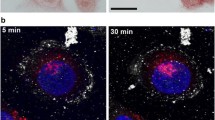
Three-dimensional information can be obtained with TEM by continuous sectioning, but both the image taking and the image analysis are time consuming. This is particularly true if the density of uptaken nanoparticles is low, since the procedure would require the analysis of many slices to obtain reliable results. For example, in Figure 4, TEM images are used to analyze the uptake of naked AuNPs in EMT cells for different co-culture times. After 30 min co-culture, only a very few AuNPs are uptaken and most TEM images show no AuNPs at all. Only by analyzing many such images we found AuNPs on the cell surface or inside the cytoplasm shown in Figure 4A and 4B. This is due to the necessary time for cells to produce and internalize the vesicles packing nanoparticles for endocytosis. This means that for short co-culture times it is difficult and time-consuming to go beyond a mere qualitative analysis.
TEM images of EMT cells for different naked AuNP co-culture times. A) co-culturing with 500 μM AuNP colloid for 30 min: only a few AuNPs can be found on the surface or inside the cytoplasm. The red square (B) marks the area where clusters are found. C) 1 h co-culture time: a larger number of endocytotic vesicles containing AuNPs is found in the cytoplasm. D) 6 h co-culture time: the number and the size of endocytotic vesicles containing AuNPs are even larger. Bar: 2 μm (A, C and D) and 200 nm (B).
X-ray imaging
Figures 5 and 6 show the main features of our x-ray micrographs in view of the quantitative analysis. Specifically, Figure 5 demonstrates that the details of 2D cultured specimens can be seen even without staining. In fact, the nucleus morphology and some subcellular details are clearly visible for EMT and HeLa 2D cell cultures- see Figures 5A and 5B. However, other organelles such as mitochondrias and vacuoles less dense and smaller than the nuclei were not fully imaged and required staining.
Transmission x-ray microimages images showing the different internalization of AuNPs by different cell lines. A) An EMT cell co-cultured with 1 mM PEG‐ coated AuNPs for 48 h. B) An EMT cell co-cultured with 500 μM naked AuNPs for 48 h. C) A HeLa cell co-cultured with 500 μM naked AuNPs for 48 h. D) A CT-26 cell co-cultured with 500 μM naked AuNPs for 48 h. (Bars: 5 μm).
Figure 6 shows that even without staining this imaging method can readily explore in detail the dissimilarities in the internalization of different AuNPs by different cell lines. Specifically, the amount of PEG coated AuNPs uptaken by EMT cells is much less than that of naked AuNPs, as seen in Figure 6A and 6B. Similar differences between these two types of AuNPs were found for all the cell lines (data not shown). For naked AuNPs, different cell lines also exhibited different responses in terms of total amount of internalized nanoparticles and nanoparticle cluster morphology. Comparing Figure 6B and 6C, it is clear that naked AuNPs are more numerous in EMT cells, form larger cluster and are distributed more evenly in cytoplasm than in HeLa cells. For comparison, we also show a similar image of naked AuNPs in CT-26 cells (Figure 6D) revealing a situation intermediate between those in EMT and HeLa cells.
These qualitative conclusions from 2D projection images can be confirmed by examining the specimens from different illumination/imaging directions as shown in Figure 7. The nanoparticle clusters appear at the nuclear membrane location of HeLa cells (Figure 7A and Additional files 1) whereas the much larger clusters inside EMT cells are distributed more uniformly throughout the cell cytoplasm. After specific staining the cell skeleton by the DAB-Ni enhancement method, we found (see Figure 7B) a close relation between the uptaken naked AuNPs and the skeletons. The high lateral resolution enabled us to detect individual naked AuNPs and to conclude from these images that no AuNPs crossed the nuclear membrane.
Transmission X-ray microimages of a 2D cultured HeLa cell. A) Co-cultured with500 μM naked AuNPs for 48h show the aggregation of AuNP clusters around the cell nucleus (Additional file 1). B) With NAB-Ni staining, AuNP clusters are imaged within the cell skeletons. Bars: 5 μm.
Additional file 1: A movie obtained from sequential projection Xray micrographs taken with 1 degree angle separation of a HeLa cell shows the aggregation of AuNP clusters around the cell nucleus (WMV 4 MB).
The morphology of 2D cultured specimens could affect the nanoparticle uptake. Therefore, we also performed tests on 3D specimens with tomographic image reconstruction. Figure 8A-C shows an example: Figure 8A is the projection image of a control EMT cell grown on a scaffold, revealing the overall cell shape. The magnified image on the right shows the nucleus (marked by arrow). Figure 8B shows similar results for a cell specimen treated with a nanoparticle colloid. Figure 8C and 8D (movie of different projection in Additional file 2) shows the results of specimen staining in revealing the detailed localization and shape of the nucleus and of the overall nanoparticle cluster distribution. Figure 8E shows the 3D tomographically reconstructed image of an EMT cell after AuNP treatment. The distribution of AuNPs and their specific location in the cell can be obtained from the rendered 3D movies (Additional file 3). It is clear that the AuNPs were not internalized in the cell nucleus. Furthermore, the cluster size distribution was quite similar to the results previously obtained on 2D cultured cell specimens.
Transmission x-ray microimages of 3D cultured EMT cells. EMT cells were grown on an OPLA scaffold. A) Cells from an untreated (without nanoparticles) specimen stained with uranium acetate. The nuclei (one of them marked by an arrow) are clearly visible. Bar: 20 μm. B) EMT cells co-cultured with 500 μM naked AuNPs for 6h. The nucleus would not be visible without staining whereas the AuNPs could be observed for unstained specimens due to their strong contrast. Bar: 5 μm. (C) Patchwork of projection micrograph for an EMT cell co-cultured with 500 μM naked AuNPs. Bar: 5 μm. (D) Single projection images like this were collected for tomographic reconstruction at 1 degree intervals with respect to the incoming x-rays (Additional file 2). Bar: 5 μm. The cell was stained with uranium acetate, targeting the lipid membrane. E) Picture of the 3D tomographic reconstruction of an EMT cell. The nanoparticle cluster distribution can be reliably extracted from the corresponding movie (Additional file 3).
Additional file 2:A movie obtained from sequential projection X-ray micrographs taken of an EMT cell grown on an OPLA scaffold with AuNPs. (WMV 8 MB)
Additional file 3: Movies from pictures of the 3D tomographic reconstruction of an EMT cell with AuNPs. (WMV 4 MB).
Figure 9 shows results for a HeLa cell 3D culture, treated with AuNP colloid and further stained. Once again, the staining procedure put in evidence the subcellular details and the nanoparticle distribution (Additional files 4 and 5 corresponding to Figure 9C and 9D). Figure 10 (and Additional file 6) shows similar results for a 3D "pellet" EMT specimen. Similar to movies associated with Figure 10B (Additional file 7), the tomographic reconstruction clearly reveals the aggregation of nanoparticles.
EMT cells prepared as the pellets. EMT cell co-cultured with 500 μM naked AuNPs and prepared as the pellets; the specimen was not stained. A) Transmission x-ray projection micrograph (Additional file 6). Bar: 5 μm. B) 3D reconstructed tomography image (Additional file 7). Bar: 5 μm. The distribution of AuNPs is shown in the corresponding movie.
Additional file 4:A movie obtained from sequential projection X-ray micrographs of a HeLa cell grown on an OPLA scaffold with AuNPs. (WMV 7 MB)
Additional file 5:Movies from pictures of the 3D tomographic reconstruction of a HeLa cell with AuNPs. (WMV 4 MB)
Additional file 6:A movie obtained from sequential projection X-ray micrographs of a pallet EMT cell with AuNPs. (WMV 7 MB)
Additional file 7:Movies from pictures of the 3D tomographic reconstruction of a pallet EMT cell with AuNPs. (WMV 7 MB)
Quantitative data
These are the core objective of our present work and the basis for the validation of our method. Figures 11 and 12 show typical results of the procedure: the size distributions of the cluster, formed by aggregation of the internalized AuNPs, for 2D and 3D cultures of EMT and HeLa cell (without staining). The distributions were obtained by analyzing 6 EMT cells and 4 HeLa cells.
These results indicate why the mere evaluation of the AuNP uptake by averaging over a large number of cells is not sufficient to understand the quantitative aspects of the phenomenon. In fact, the above figures reveal substantial differences between different types of cells and even between different cells of the same type.
We see indeed distributions with different peaks and different spreads. Overall, the data for EMT cells suggest a size distribution peak around 140 nm for EMT cells and around 30 nm for HeLa cells. These cluster distribution differences are statistically significant. Thus, our approach made possible a quantitative evaluation of AuNP cluster distributions at the individual cell level, yielding relevant information for the intracellular uptake mechanism that cannot be delivered by cell-averaging procedures.
In addition, our microimages also revealed interesting qualitative differences between EMT and HeLa cells. We see in fact from Figures 11 and 12 that the clusters are concentrated near the nucleus for HeLa cells, whereas they are more uniformly distributed for EMT cells. By measuring the location of the AuNP clusters with respect to the nucleus membrane from 3D images such as Figure 7A, we could determine that the clusters were internalized within a very narrow region, ~1 ± 0.5 μm, outside the HeLa cell nuclear membrane.
The difference of the AuNP internalization process between cell lines can be explained by differences in the biophysical mechanisms: the endocytosis depends on the cell membrane properties, which can be largely different between epithelial, endothelial, cubic, circulating, etc. phenotypes. The different size of the clusters encompassed by endosomes could also result in different intracellular transportation mechanisms of the AuNP clusters. One must also consider that biochemical environment will play a major role in this process, with pH, fluid pressure, and interstitial homeostasis modulating the size and the number of formed vesicles. Further studies on the details of the dynamics of these processes with respect to the size of the clusters are underway.
Figure 13 emphasizes another important quantitative issue: the difference between 2D and 3D analysis. The figure shows four x-ray micrographs from 3D pellet specimens of EMT cells. These projection images are from large angular sets (Additional file 8) from which tomographically reconstructed pictures were obtained. The analysis of such pictures yielded the cluster size distribution also shown in Figure 13.
Quantitative uptake analysis of EMT plellets by 3D images. A) Internalized cluster size distribution from four x-ray transmission micrographs (B) of 3Dal pellet specimens of EMT cells. Bars: 5 μm. (Additional file 8).
Additional file 8:A movie obtained from sequential projection X-ray micrographs of specimen S3 (pallet EMT cells) in Figure 13 taken at 1 degree intervals. (WMV 7 MB)
It is clear that such a distribution is substantially different from the corresponding 2D results, Figure 11. This means that the reliable extraction of data on uptaken nanoparticles must include the analysis of 3D specimens, made possible in our approach by the combination of x-ray microscopy and tomographic reconstruction.
On the qualitative side, 3D data on pellet specimens corroborated the information from 2D cultured specimens. Specifically, they confirmed that clusters inside EMT cells are substantially larger and more uniformly distributed than inside HeLa cells, even when the cells are prepared in 3D.