Abstract
Type 1 diabetes (T1D) patients with low genetic risk scores (GRS) may be non-autoimmune or autoimmune mediated by other genetic loci. The T1D-GRS2 provides us an opportunity to look into the genetic architecture of these patients. A total of 18,949 European individuals were included in this study, including 6599 T1D cases and 12,323 controls. 957 (14.5%) T1D patients were identified with low GRS (GRS < 8.43). The genome-wide association study on these patients identified 41 unreported loci. Two loci with common variants and 39 loci with rare variants were identified in this study. This study identified common SNPs associated with both low GRS T1D and expression levels of the interferon-α-induced MNDA gene, indicating the role of viral infection in T1D. Interestingly, 16 of the 41 unreported loci have been linked to autism spectrum disorder (ASD) by previous studies, suggesting that genes residing at these loci may underlie both T1D and autism.
Similar content being viewed by others

Introduction
Type 1 diabetes (T1D) has been traditionally recognized as an autoimmune disease, and its molecular immunological mechanisms have been corroborated by the discovery of numerous autoimmune disease genes underlying the genetic susceptibility of T1D1,2,3. As we showed in a recent study4, the genetic risk of T1D can be predicted by genome-wide DNA variants generated using a global polygenic risk scoring (PRS) approach5. In contrast, Sharp et al. developed a specific genetic risk scoring (GRS) system for T1D, (T1D-GRS2), using 67 single nucleotide polymorphisms (SNPs) from known autoimmune loci associated with T1D, while haplotypic effects and interactions of common human leukocyte antigen (HLA) DR-DQ haplotypes, conferring the primary effects to T1D susceptibility1,6,7,8, were also taken into account9. In contrast to global PRS scoring using genome-wide DNA markers, T1D-GRS2 uses a small set of T1D genetic markers. Among the 67 SNPs, 35 are from the major T1D susceptibility loci, i.e., the HLA region accounting for about 50% of the genetic susceptibility in the European population1. The other 32 SNPs are from 31 non-HLA T1D susceptibility loci.
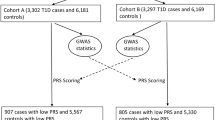
T1D is a complex and heterogeneous phenotype. A minor proportion (~5–10%) of Caucasian patients diagnosed with T1D have non-autoimmune pathogenesis, i.e., T1bD10. Moreover, there are also autoimmune patients (e.g., with islet cell auto-antibodies), but with low-risk genotypes of known T1D genes, e.g., protective HLA haplotypes11. The PRS approach using genome-wide DNA markers represents primarily autoimmune T1D4, whereas low PRS suggests non-autoimmune mechanisms may be involved. As expected, a number of loci identified in our analysis have been reported of association with obesity-related traits by previous GWA studies4. Our gene-based association study in patients with low PRS identified the Notch ligand Delta-like 1 gene (DLL1)12, involved in impaired glucose tolerance and reduced insulin secretion13. In contrast, low GRS may also include autoimmune patients with undetermined genetic mechanisms. Thus, patients with T1D and low GRS may have their disease susceptibility conferred by other unknown genetic loci. Due to these potentially distinct biological mechanisms underlying T1D captured by PRS vs. GRS, respectively, the T1D-GRS2 scoring system provides us a unique opportunity to identify patients with low GRS and consequently enables us to look into the genetic architecture of this group of patients.
Results
GRS scores
As shown by our ROC analysis, the GRS scores for T1D prediction have the performance of the Area Under the ROC Curve (AUC) = 0.866. The cutoff of GRS = 8.43 has the maximum Matthews correlation coefficient (MCC) of 0.580, with the sensitivity to identify T1D patients (true positive rate, TPR) of 0.855 and the specificity to identify individuals without T1D (true negative rate, TNR) of 0.719 (Supplementary Data 2). We identified 957 (14.5%) T1D patients with GRS < 8.43.
In this study, the GRS scores are significantly correlated with the PRS scores (P < 1E − 200), with Pearson’s r = 0.305 and 0.331, respectively in the two PRS cohorts, as we reported4. Among the 6599 T1D cases, 4314 (65.4%) cases have both high GRS and high PRS scores; 381 (5.8%) cases have both low GRS and low PRS scores; 1328 (20.1%) cases have high GRS and low PRS; and 576 (8.7%) cases have low GRS and high PRS.
GWAS of T1D patients with low GRS
Nine hundred and fifty-seven T1D patients (474 males and 483 females) were identified with low GRS. The HLA loci contributed significantly to this GRS classification (Table 1), while the HLA-DQ locus contributes more to the classification (beta = 0.489) than the combined effects of other HLA loci (beta = 0.308). The GWAS results of patients with low T1D GRS vs. all non-diabetes controls (Fig. 1a) were compared with that of all T1D patients vs. all non-diabetes controls (Fig. 1b). As expected, the majority of SNPs showing genome-wide significance in low T1D GRS were also significant in the overall T1D cohort (Supplementary Data 3), which is in kee** with our original hypothesis that low GRS cases may also be driven by autoimmune mechanisms. However, the potential undetermined genetic mechanisms of the low T1D GRS patients are highlighted by 82 single nucleotide variants (SNV) from 47 non-HLA genetic loci with genome-wide significance in the low GRS T1D cases only, but not significant in the overall T1D cohort (Supplementary Data 4). The minor alleles of 81 out of the 82 SNVs are predisposing, except the SNP rs62425513 at the THEMIS locus. Without exception, however, the genetic effects represented by these SNPs in the low GRS T1D cases are all greater than the effects observed in the overall T1D cohort, where each SNP’s OR in the overall T1D cohort fell outside the 95% confidence interval (95% CI) in the low GRS T1D cohort, although 11 SNVs from 8 loci have the two-tailed P values of heterogeneity test >0.05.
Unreported loci with common variants associated with low GRS T1D
Among the 82 genome-wide significant SNPs, 19 common SNPs from 5 independent genetic loci have minor allele frequencies (MAF) > 0.050. 16 common SNPs from 2 genetic loci have MAF in the range of 0.304 to 0.462. (1) The locus at chr1q23.1 (Fig. 2) harbors several coding genes, including the myeloid cell nuclear differentiation antigen gene (MNDA), encoding an interferon-inducible gene, and the pyrin and HIN domain family member 1 gene (PYHIN1), encoding an interferon-inducible gene. The strongest association signal in low T1D GRS (Fig. 2a), which is lack significance in all T1D patients (Fig. 2b), is tagged by the SNP rs857786 upstream of MNDA, with OR (95% CI) = 1.322 (1.203, 1.452), P = 6.44E − 09, representing a strong effect size for a common SNP. (2) The association signal in low T1D GRS [Fig. 3a, lack of significance in all T1D patients (Fig. 3b)], tagged by the SNP rs13147255 with OR(95% CI) = 1.318(1.198,1.449), P = 1.42E − 08, at the chr4q28.1 locus, resides between the long intergenic non-protein coding RNA 2516 gene (LINC02516) and the ankyrin repeat domain 50 genes (ANKRD50).
Unreported loci with rare variants associated with low GRS T1D
Among the 42 independent loci with rare variants (MAF < 0.050), one locus (i.e., the PGM1 locus) has been reported of association with T1D by previous GWAS studies (Supplementary Data 5, GWAS Catalog, https://www.ebi.ac.uk/gwas/) and two recent large-scale T1D GWAS14,15, thus was not taken as an unreported locus. In addition, the DOK6 locus identified in this study, led by the SNV rs146427450 with OR(95% CI) = 2.858(2.076, 3.935), P = 1.22E − 10, is close to (~227 kb) the CD226 locus identified by the studies by Robertson et al.15 and Crouch et al.14 Another locus tagged by the SNV rs148505224 at CFTR with OR(95% CI) = 3.107(2.090, 4.619), P = 2.09E − 08, is close to the ASZ1 locus recently identified by Crouch et al.14 Besides these 3 loci and the above loci with common genetic variants associated with low GRS T1D, we uncovered 39 unreported loci with variants in the low to rare frequency range (MAF ≤ 3.92% in this study) associated with low GRS T1D (Supplementary Data 4).
Discussion
Five loci with common variants were identified of association with low GRS T1D in this study with genome-wide significance. Besides the above 2 loci with common variants, 3 SNPs from 3 different loci (MIR4278/MIR4454, THEMIS, MSRB3) have also common SNPs with minor allele frequencies (MAF) > 0.050 associated with low GRS T1D (Figs. 4a, 5a, 6a), but not the general T1D cases (Figs. 4b, 5b, 6b). However, as shown in Figs. 4–6, different SNPs at each loci are associated with the general T1D cases, therefore these 3 loci were not taken as unreported loci specifically associated with low GRS T1D in this study. In addition, the THEMIS locus tagged by the SNP rs62425513 with OR (95% CI) = 0.673(0.584,0.775), P = 3.76E − 08, was identified of T1D association with FDR < 0.01 by Robertson, et al.15. Interestingly, each of these 3 loci has been identified of association with obesity-related traits or waist-hip ratio by the previous GWASs (Supplementary Data 5). The THEMIS locus was also reported of association with celiac disease by the previous GWAS16.
The unreported T1D genetic locus MNDA is involved in interferon signaling. At the OR6N2/MNDA/PYHIN1 locus, the strongest association signal rs857786 with OR(95% CI) = 1.322(1.203,1.452), P = 6.44E − 09, at the 5′-upstream of MNDA, is also associated with the gene expression of MNDA in whole blood (P = 9.9e − 14), according to the expression quantitative trait loci (eQTLs) data of the GTEx Project (https://www.gtexportal.org/)17. The protein encoded by MNDA is expressed specifically in hematopoietic cells, and upregulated by interferon-α18. Viral infections have been suggested as a possible trigger of T1D, although the evidence remains controversial19,20. Interferon-α is a potential link of viral infection and autoimmunity in T1D21. In our study, significant association from the interferon-α-induced MNDA locus is only seen in low GRS cases, but not in overall T1D cases (Fig. 2). This may imply a plausible explanation about the pathogenesis of low GRS T1D patients, i.e., that viral infection contributes to the T1D pathogenesis in these patients despite the low overall genetic risk. In addition to our study, a previous study has shown an association of this locus with monocyte chemoattractant protein-1 levels22.
The unreported T1D genetic locus LINC02516/ANKRD50 is involved in retromer function. ANKRD50 has been demonstrated of essential role in the function of retromer and the endocytic recycling23. The protein encoded by ANKRD50 is an essential component for the retromer function23. The retromer mediates the retrograde transport from the endosome to the Golgi24. The retromer protein VPS35 which mediates the retromer cargo selection has been shown to be associated with T2D in a previous GWAS study25. The gene encoding a receptor of VPS35, the sortilin related VPS10 domain-containing receptor 1 gene (SorCS1), has also been reported in association with glycemic control in T1D26 and insulin secretion in T2D27.
Association between autism and T1D has been reported previously28. Interestingly, 15 of the 39 unreported loci identified of genome-wide significance in this study have been reported to harbor variants predisposing to autism or autism spectrum disorder (ASD) according to the HGMD database (http://www.hgmd.cf.ac.uk) (Supplementary Data 4), and 2 of these loci have been previously reported of association with ASD by the previous GWASs29,30. Six genes at these loci are expressed at the cell synapse (cellular_component GO:0045202), including ankyrin 3 (ANK3), cell cycle associated protein 1 (CAPRIN1), cadherin 8 (CDH8), fibroblast growth factor receptor 2 (FGFR2), olfactomedin 3 (OLFM3), and prion protein (PRNP). Mechanisms of neural control of the endocrine pancreas31 mediated by these genes are therefore highlighted. Besides the 15 loci linked to autism or ASD, two other loci with rare variants associated with low GRS T1D encode synapse-expressed genes, abhydrolase domain containing 17B (ABHD17B) and leucine-rich repeat and fibronectin type III domain containing 3 (LRFN3). It is also worth mentioning that, at the OR6N2/MNDA/PYHIN1 locus with common SNPs associated with low GRS T1D, the pyrin and HIN domain family member 1 gene (PYHIN1) whose expression is also interferon-induced has also been linked to autism and ASD by the previous study32.
The SNV rs148505224 associated with low GRS T1D maps to the CF transmembrane conductance regulator (CFTR) gene, i.e., the gene mutated in cystic fibrosis(CF) and a cause of pancreatitis. As this CFTR locus is close to the ASZ1 locus recently identified by Crouch et al.14, it is not counted as an unreported locus identified in this study. The chr4p15.2 locus encoding the peroxisome proliferator-activated receptor gamma (PPAR-γ) coactivator 1 alpha gene (PPARGC1A), which has been shown to important roles in energy homeostasis33. Besides the 3 loci with common SNPs associated with waist-to-hip ratio or obesity-related traits, 8 genetic regions with rare variants demonstrating an association with low GRS T1D have been previously reported in association with body mass index (BMI), weight, waist-hip ratio, or obesity-related traits (Supplementary Data 5).
In addition to the MNDA locus, two loci have been reported of association with severe influenza A (H1N1) infection, including the locus at chr2q14.1 encoding the dipeptidyl peptidase like 10 genes (DPP10) and the locus at chr19q13.12 containing the nuclear factor kappa B (NF-κB) inhibitor delta gene (NFKBID)34. NFKBID is critical for B cell development as shown in mouse model35. In addition to the association of THEMIS with celiac disease, two loci with rare variants have been reported of association with autoimmune diseases, i.e., the long intergenic non-protein coding RNA 1967 (LINC01967) and C-X9-C motif containing 1 (CMC1) locus at chr3p24.1 has been reported to be associated with multiple sclerosis36, and the small nucleolar RNA, C/D box 3F (SNORD3F)/leucine zipper tumor suppressor 1 (LZTS1) antisense RNA 1 (LZTS1-AS1) locus at chr8p21.3 which has been reported to be associated with rheumatoid arthritis37.
In conclusion, this study identified 41 unreported loci associated with low GRS T1D. In addition to our previous study on low PRS T1D which identified new non-autoimmune T1D loci4, this study identified common genetic variants at two loci related to interferon signaling involved in viral infection and retromer function, respectively. The role of viral infection in low GRS T1D is supported by the common SNPs associated with low GRS T1D inferring considerable effect size (OR~1.32). Likewise, unreported loci related to pancreatitis, BMI, and obesity were also uncovered. 16 of 41 loci have been previously linked to autism or ASD38,39 (http://www.hgmd.cf.ac.uk). Accordingly, this study highlights a number of genes that may mediate shared molecular mechanisms of ASD and T1D. From the molecular genetics aspect, these rare variants may suggest a possibility of rare syndromic types of disease with clinical characterizations of both ASD and T1D not previously identified. Additional studies are needed to confirm our hypothesis and preliminary results, that patients with both ASD and T1D diagnosis may share common genetic factors. Patients with low T1D GRS may also be autoimmune, i.e., T1aD with undetermined genetic mechanisms, or non-autoimmune, e.g., T1bD. Among the genetic loci identified in this study, 3 loci with common SNPs and 8 genetic regions with rare variants have been previously reported in association with BMI, waist-to-hip ratio, or obesity-related traits, suggesting T1bD. It is also worth pointing out that these reported variants may still be autoimmune T1D loci, suggested by the two loci reported of association with autoimmune diseases, and the two loci associated with severe influenza A infection. Patients with low T1D GRS, whether they are T1aD or not, may have their disease susceptibility conferred by these loci with inflated effect sizes in this subgroup of patients.
Methods
Subjects
A total of 18,949 European individuals were included in this study, including 6599 T1D cases and 12,323 controls. The T1D cases were from Montreal Children’s Hospital and the Children’s Hospital of Philadelphia (CHOP)3, The Diabetes Control and Complications Trial – Epidemiology of Diabetes Interventions and Complications (DCCT-EDIC) cohort (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000086.v2.p1), and the Type 1 Diabetes Genetics Consortium (T1DGC, http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000180.v1.p1), respectively. Informed consent was obtained from each of the relevant cohorts/studies. The genoty** was done by the Illumina Genoty** BeadChips with at least 550,000 SNPs genotyped. More details of these research subjects are previously described3,40. All the research subjects have been confirmed of European ancestry by principal component analysis (PCA) with genomic DNA markers. Genome-wide imputation was done by the TOPMed Imputation Server (https://imputation.biodatacatalyst.nhlbi.nih.gov) with the TOPMed (Version R2 on GRC38) Reference Panel.
GRS scoring
The GRS scoring was based on the method developed by Sharp et al.9. To acquire the genotype information of all the T1D-GRS2 SNPs (67 SNP markers, Supplementary Data 1), the HLA region was additionally imputed by the SNP2HLA software41. The overlapped SNPs covered across the imputation methods were highly consistent. Consequently, the GRS scores were assessed for their predictive performance by AUC. The GRS cutoff for low GRS vs. high GRS was determined by the maximum MCC, which represents a balanced measure of sensitivity and specificity.
Statistics and reproducibility
We tested 104,689,647 autosomal SNV with quality filters of R2 ≥ 0.3 for the genetic association, using 12,323 controls (6665 males and 5658 females) and 6599 T1D cases, including 957 T1D patients (474 males and 483 females) with low GRS. Genetic association tests were performed using PLINK1.9 software42, conditioned on sex, and corrected by the first 10 principal components (PC) of population structure analysis. Genome-wide significance was defined as P < 5 × 10−8. The Manhattan plots were done using the SNPEVG software43. Genetic association signals within each locus were plotted by LocusZoom44. The genetic association of T1D patients with low GRS was compared with that of the general T1D patients by heterogeneity Z test45. We defined each locus by r2 > 0.5 from lead SNPs46.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Summary statistics are presented in Supplementary Data 4 and are available at the NHGRI-EBI GWAS catalog (GCP ID: GCP000182, https://www.ebi.ac.uk/gwas). The original genoty** data are available from dbGAP. All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Todd, J. A., Bell, J. I. & McDevitt, H. O. HLA-DQ[beta] gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329, 599–604 (1987).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Hakonarson, H. et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 448, 591–594 (2007).
Qu, J. et al. Genetics of low polygenic risk score type 1 diabetes patients: rare variants in 22 novel loci. medRxiv (2020).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. GigaScience 8, giz082 (2019).
Todd, J. A. Genetic analysis of type 1 diabetes using whole genome approaches. PNAS 92, 8560–8565 (1995).
Noble, J. A. et al. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am. J. Hum. Genet. 59, 1134–1148 (1996).
she, J.-X. Susceptibility to type I diabetes: HLA-DQ and DR revisited. Immunol. Today 17, 323 (1996).
Sharp, S. A. et al. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care 42, 200–207 (2019).
Leslie, R. D., Atkinson, M. A. & Notkins, A. L. Autoantigens IA-2 and GAD in Type I (insulin-dependent) diabetes. Diabetologia 42, 3–14 (1999).
Noble, J. A. & Valdes, A. M. Genetics of the HLA region in the prediction of type 1 diabetes. Curr. Diabetes Rep. 11, 533 (2011).
Qu, J. et al. Association of DLL1 with type 1 diabetes in patients characterized by low polygenic risk score. Metabolism 114, 154418 (2020).
Rubey, M. et al. DLL1- and DLL4-mediated notch signaling is essential for adult pancreatic islet homeostasis. Diabetes 69, 915–926 (2020).
Crouch D. J. et al. Enhanced genetic analysis of type 1 diabetes by selecting variants on both effect size and significance, and by integration with autoimmune thyroid disease. bioRxiv (2021).
Robertson C. C. et al. Fine-map**, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. bioRxiv (2020).
Coleman, C. et al. Common polygenic variation in coeliac disease and confirmation of ZNF335 and NIFA as disease susceptibility loci. Eur. J. Hum. Genet. 24, 291–297 (2016).
Lonsdale, J. et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Briggs, R. C. et al. The human myeloid cell nuclear differentiation antigen gene is one of at least two related interferon-inducible genes located on chromosome 1q that are expressed specifically in hematopoietic cells. Blood 83, 2153–2162 (1994).
Van der Werf, N., Kroese, F. G., Rozing, J. & Hillebrands, J. L. Viral infections as potential triggers of type 1 diabetes. Diabetes/Metab. Res. Rev. 23, 169–183 (2007).
Filippi, C. M. & von Herrath, M. G. Viral trigger for type 1 diabetes: pros and cons. Diabetes 57, 2863–2871 (2008).
Devendra, D. & Eisenbarth, G. Interferon alpha—a potential link in the pathogenesis of viral-induced type 1 diabetes and autoimmunity. Clin. Immunol. 111, 225–233 (2004).
Ahola-Olli, A. V. et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am. J. Hum. Genet. 100, 40–50 (2017).
Kvainickas, A. et al. Retromer- and WASH-dependent sorting of nutrient transporters requires a multivalent interaction network with ANKRD50. J. Cell Sci. 130, 382–395 (2017).
Trousdale, C. & Kim, K. Retromer: structure, function, and roles in mammalian disease. Eur. J. Cell Biol. 94, 513–521 (2015).
Kooner, J. S. et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 43, 984–989 (2011).
Paterson, A. D. et al. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes 59, 539–549 (2010).
Goodarzi, M. O. et al. SORCS1: a novel human type 2 diabetes susceptibility gene suggested by the mouse. Diabetes 56, 1922–1929 (2007).
Freeman, S. J., Roberts, W. & Daneman, D. Type 1 diabetes and autism: is there a link? Diabetes Care 28, 925–926 (2005).
Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Rodriguez-Diaz, R. & Caicedo, A. Neural control of the endocrine pancreas. Best. Pract. Res. Clin. Endocrinol. Metab. 28, 745–756 (2014).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
Esterbauer, H., Oberkofler, H., Krempler, F. & Patsch, W. Human peroxisome proliferator activated receptor gamma coactivator 1 (PPARGC1) gene: cDNA sequence, genomic organization, chromosomal localization, and tissue expression. Genomics 62, 98–102 (1999).
Garcia-Etxebarria, K. et al. No major host genetic risk factor contributed to A(H1N1)2009 Influenza severity. PLoS ONE 10, e0135983 (2015).
Touma, M. et al. Impaired B cell development and function in the absence of IkappaBNS. J. Immunol. 187, 3942–3952 (2011).
International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019).
Saad, M. N., Mabrouk, M. S., Eldeib, A. M. & Shaker, O. G. Studying the effects of haplotype partitioning methods on the RA-associated genomic results from the North American Rheumatoid Arthritis Consortium (NARAC) dataset. J. Adv. Res. 18, 113–126 (2019).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 179, 1469–1482.e1411 (2019).
The Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21 (2017).
Bradfield, J. P. et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 7, e1002293 (2011).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PloS ONE 8, e64683 (2013).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, s13742-13015–10047-13748 (2015).
Wang, S., Dvorkin, D. & Da, Y. SNPEVG: a graphical tool for GWAS graphing with mouse clicks. BMC Bioinforma. 13, 319 (2012).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Altman, D. G. & Bland, J. M. Interaction revisited: the difference between two estimates. BMJ 326, 219 (2003).
Pers, T. H. et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 6, 1–9 (2015).
Acknowledgements
Dr. Hakon Hakonarson is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. The study was supported by Institutional Development Funds from the Children’s Hospital of Philadelphia to the Center for Applied Genomics and The Children’s Hospital of Philadelphia Endowed Chair in Genomic Research to H.H.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.H., C.P., and H.Q.; literature search: H.Q. and L.M.; figures and tables: H.Q., and J.Q.; data analysis: H.Q., J.Q., and J.B.; data interpretation: H.Q., J.Q., L.M., J.G., X.C., M.M., J.L., J.J.C., J.D.R., P.S., C.P., and H.H.; original draft writing: H.Q., J.Q., and H.H.; review and revision: H.Q., C.P., and H.H.; supervision: H.H. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Chiea Chuen Khor and George Inglis.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qu, HQ., Qu, J., Bradfield, J. et al. Genetic architecture of type 1 diabetes with low genetic risk score informed by 41 unreported loci. Commun Biol 4, 908 (2021). https://doi.org/10.1038/s42003-021-02368-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02368-8
- Springer Nature Limited