Abstract
Growth of plants in soil inoculated with plant growth promoting bacteria (PGPB) producing 1-aminocyclopropane-1-carboxylate (ACC) deaminase or expression of the corresponding acdS gene in transgenic lines reduces the decline in shoot length, shoot weight and photosynthetic capacity triggered by salt stress in Camelina sativa. Reducing the levels of ethylene attenuated the salt stress response as inferred from decreases in the expression of genes involved in development, senescence, chlorosis and leaf abscission that are highly induced by salt to levels that may otherwise have a negative effect on plant growth and productivity. Growing plants in soil treated with Pseudomonas migulae 8R6 negatively affected ethylene signaling, auxin and JA biosynthesis and signalling, but had a positive effect on the regulation of genes involved in GA signaling. In plants expressing acdS, the expression of the genes involved in auxin signalling was positively affected, while the expression of genes involved in cytokinin degradation and ethylene biosynthesis were negatively affected. Moreover, fine-tuning of ABA signaling appears to result from the application of ACC deaminase in response to salt treatment. Moderate expression of acdS under the control of the root specific rolD promoter or growing plants in soil treated with P. migulae 8R6 were more effective in reducing the expression of the genes involved in ethylene production and/or signaling than expression of acdS under the more active Cauliflower Mosaic Virus 35S promoter.
Similar content being viewed by others
Introduction
The ability of Camelina sativa (camelina) to grow on marginal lands has piqued interest in its development as an industrial oilseed crop for biofuels, bio-lubricants and animal feed1,2. C. sativa is an allopolyploid that arose from a genome triplication event3 resulting in a 750 Mb genome encoding 89,418 proteins4. The genome remains highly undifferentiated with little fractionation bias; this presents significant challenges for breeding and genetic manipulation4,5,6, necessitating alternate strategies for trait improvement.
Accumulation of salts at or near the soil surface is a widespread agricultural problem that causes cellular dehydration and ion toxicity in plants7,8. These have deleterious effects on biochemical reactions resulting in plasmolysis, accumulation or reduction of specific secondary metabolites, nutrient imbalance, and production of reactive oxygen species, all of which can interfere with photosynthesis9. The consequences of these physiological changes are inhibition of seed germination, reduction in plant growth and vigor, changes to flowering time, and reduction in seed production7,10. While plants may respond somewhat differently to various stresses, nearly all respond by producing ethylene. Following the onset of stress, a small increase in ethylene production induces the transcription of defense/stress response genes. A subsequent and larger burst of ethylene is generated by persistent stress that acts as a signal to initiate processes, such as senescence, chlorosis and abscission, all of which inhibit plant growth11,12. The synthesis of stress ethylene can be reduced by lowering the level of the ethylene precursor, 1-aminocyclopropane-1-carboxylate (ACC), by applying the enzyme ACC deaminase either in the form of bacteria producing the enzyme or by introducing the bacterial acdS gene into the plant. These approaches significantly reduce the extent of inhibition on plant growth and development that normally accrues from stress13,14,15,16. Canola (Brassica napus) grown in soil treated with a strain of Pseudomonas putida producing ACC deaminase exhibited up-regulation of genes encoding auxin response factors and down-regulation of stress response genes11. Furthermore, transgenic B. napus expressing acdS performed better under salinity stress compared to non-transgenic plants17. Similarly, C. sativa lines expressing acdS or treated with bacteria producing ACC deaminase exhibited increased salinity tolerance16; this dramatically affected the transcriptional response in the roots of these plants under salt stress18. Here, we examined how expression of acdS in transgenic C. sativa under the control of broadly constitutive (CaMV 35S) or root-specific (rolD) promoters, or treatment with plant growth promoting bacteria (PGPB) producing ACC deaminase affected the transcription of genes in vegetative tissues in response to salt stress.
Materials and methods
Bacterial strains
Two PGPB root endophytes that produce ACC deaminase, Pseudomonas migulae 8R6 and Pseudomonas fluorescens YsS619, that were previously shown to increase salinity tolerance in C. sativa16 were used in this study. Two derived acdS mutant strains, 8R6M and YsS6M, constructed previously by insertion of a transposon containing the tetracycline resistance gene at position 237 in the P. migulae 8R6 acdS gene and at position 323 in the P. fluorescens YsS6 acdS gene13 were also examined. The ACC deaminase activity for these bacteria was determined previously13. The ACC deaminase activity of the wild-type P. fluorescens YsS6 was 12.5 mmol mg−1 h−1, while the activity of the corresponding acdS mutant was 0.11 mmol mg−1 h−1. The ACC deaminase activity of the wild-type P. migulae 8R6 was 10.9 mmol mg−1 h−1 and the activity of the corresponding acdS mutant was 0.03 mmol mg−1 h−1.
Transgenic C. sativa lines expressing acdS
Camelina sativa cv. DH55 lines expressing the Pseudomonas sp. UW420 acdS gene under the control of either the double cauliflower mosaic virus (CaMV) 35S promoter or the rolD promoter from Agrobacterium rhizogenes were constructed previously16. The P. sp. UW4 ACC deaminase is 99% identical (amino acid level) to that from P. fluorescens YsS6 and 98% identical to that from P. migulae 8R616. Individual, single copy lines in which the expression of the P. sp. UW4 acdS gene was verified using droplet digital PCR16 and RNA-Seq data18 were used.
PGPB and salt treatment
Bacteria were cultured for 24 h in tryptic soy broth (TSB) containing 100 μg ml−1 ampicillin for wild-type strains or 100 μg ml−1 ampicillin and 10 μg ml−1 tetracycline for the acdS mutant strains21. Bacterial cultures were centrifuged at 4000×g and washed three times with 0.03 M MgSO4 prior to re-suspending to an OD600nm = 0.50 ± 0.02 in 0.03 M MgSO4 (approximately 5.0 × 108 bacteria/ml). Soil was inoculated with either 2 ml of PGPB or 2 ml of 0.03 M MgSO4 (control) at the time of sowing and again 1 week after sowing.
Camelina sativa cv. DH55 seeds were sown in soil-less potting mixture22 in 10 × 10 × 8 cm square pots. All materials were available locally, except fritted trace elements (Frit Industries Inc., Ozark, USA). NaCl solutions at 192 and 213 mM were prepared to obtain solutions with electrical conductivities (EC) of 15 dS m−1 and EC 20 dS m−1 at 20 °C, respectively. Tap water (50 ml; EC 248 µS m−1) was applied daily to each pot and replaced with 50 ml of saline solution 20 days after sowing. The accumulation of salt in the pots was controlled by draining and the EC of the drained water was measured weekly. Plants were grown in a controlled growth cabinet (16 h light/8 h dark; 20/17 °C) supplemented with halogen lights. Shoot material was harvested 20 days after the initial salt treatment just as plants began to flower, at which time the length and dry weights were recorded. Shoot length was measured from axis of the first true leaf petiole with the stem to the tip of the shoot and recorded to the nearest millimeter. For dry weight, samples were placed in pre-weighed, dry beakers and then dried at 80 °C for 48 h in a drying oven and weight recorded to the nearest milligram.
Element, chlorophyll and ethylene measurement
The quantity of elements, chlorophyll and ethylene were determined in shoots of 33-day-old plants; 13 days after initial salt treatment and before plants exhibited severe symptoms of salt stress18. Samples were dried at 80 °C for 48 h and ground using liquid nitrogen. Sulphur (S), copper (Cu), boron (B), calcium (Ca), iron (Fe), potassium (K), magnesium (Mg), manganese (Mn), sodium (Na), phosphorus (P), and zinc (Zn) content were determined by Agvise Laboratories Inc. (Benson, MN, USA). The method employed involved digestion with nitric acid/hydrogen peroxide and determination of elements using inductively coupled plasma mass spectrometry on a Perkin Elmer 5400 analyzer (Perkin Elmer, Guelph, Canada) according to Wu et al.23.
Chlorophyll content was measured using a chlorophyll meter (Apogee Instruments Inc., Logan, UT, USA) and photosynthesis yield (Fv/Fm) using a FV/FM meter (ADC Bioscientific Ltd., UK)12.
For ethylene measurement, 4 g (wet weight) of vegetative tissue was placed in 40 ml glass vials. The vials were sealed with rubber stoppers and placed at 20 °C for 6 h. 20 ml of gas was removed with a syringe and injected into a pre-evacuated Exetainer soda glass vial paired with a cap constructed with both a PTFE/silicone and a chlorobutyl septum (Labco Ltd., Ceredigion, UK). Ethylene was separated by gas chromatography (Bruker 450 GC, Bruker Biosciences Corporation, USA) and its concentration was calculated by comparing the peak area and peak length to standard peaks generated with pre-determined ethylene concentrations. Ethylene production was reported as parts per million ethylene/g fresh weight/hour. The measurements were repeated with three independent biological replicates.
RNA sequencing and data analysis
Total RNA was extracted from shoot tissue of three biological replicates for each of the control, 35S::acdS, rolD::acdS and P. migulae 8R6 treated lines (12 samples in total) from 28-day-old plants (21 days after 15 dS m−1 salt treatment began and before plants started to bolt) using an RNeasy Plant Mini Kit (Qiagen Inc.)12,18. The cDNA libraries were prepared using the TruSeq Stranded mRNA and Total RNA Library Prep kits with TruSeq LT adaptors (Illumina Inc.). The tagged libraries were sequenced using a HiSeq 2500 (Illumina Inc.)12,18.
The short-read sequence data from the 12 libraries were deposited in the NCBI GEO database (GSE132600). Trimmomatic24 was used to discard low-quality reads, to remove adaptor sequences and low quality nucleotides at the beginning or end of the read (PHRED33 quality score of less than 3), and to discard short reads (under 21 nt). The retained high-quality reads were mapped to the C. sativa reference genome using STAR RNA-seq aligner25 and transcripts available in the NCBI C. sativa release 100 (JFZQ00000000.14). Expression levels for each gene were measured as counts26,27. Normalization for library size was performed for each gene by dividing the counts by the library size as described28 to yield counts per million (CPM). Further normalization was performed using DESeq to approximate a negative binomial distribution. The differential expression analysis of digital gene expression data software (edgeR) was used to calculate differences in expression between libraries26. The Biological Coefficient of Variation (BCV) value was set to 2 according to the software's instructions. Expression changes were declared to be significant if the multiple test corrected p value and the false discovery rate (FDR) were ≤ 0.05 and the absolute value of log 2 CPM was higher than 1. MAPMAN software version 3.5.1R2 (http://mapman.gabipd.org/) was used to assign gene ontology (GO) terms to unigenes based on molecular function, biological processes and cellular compartment29. Cluster analysis was done using JMP software version 15 (https://www.jmp.com/en_ca/home.html).
Quantitative droplet digital PCR (ddPCR) analysis
Quantitative ddPCR was used to confirm the expression profiles of a set of genes as determined by the RNA-Seq analysis. Total RNA was extracted from shoots of 3 independent biological replicates (control, 35S::acdS, rolD::acdS and P. migulae 8R6 treatment) using the RNeasy Plant Mini kit (Qiagen) and cDNA synthesized using the SuperScript III First-Strand Synthesis kit (Thermo Fisher Scientific). One ng of cDNA was used as template in each PCR reaction mixture with 2× Supermix (Bio-Rad Laboratories Inc.), 500 nM of each primer and 250 nM of each probe in a final volume of 20 µl. The sequences of the primers and probes are provided in Supplemental Data 1. Data were analyzed using Quanta-Soft version 1.7.4.0917 (Bio-Rad Laboratories Inc.) and the relative ratio of the candidate gene expression was calculated relative to the expression of the actin reference gene by plotting the concentration of FAM- over the HEX-labelled probe according to the Bio-Rad ddPCR guide12,18.
Statistical analysis
Plant growth measurements were expressed as the mean ± standard error for each treatment. Significant differences between treatments were determined by variance analysis (ANOVA) with a p value of ≤ 0.05 and pair-wise comparisons were conducted using the Tukey's Studentized Range (HSD) test using SAS 9.3 (TS1M2)12,16,18.
Results
Ethylene, plant growth, photosynthetic capacity, and element composition
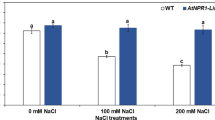
Expression of acdS or treatment of soil with P. migulae 8R6 had only a marginal effect on the levels of ethylene in vegetative tissue in the absence of salt (Fig. 1). Overall, levels of ethylene were lower in all lines sampled at 13 days after the application of salt; however, the levels were significantly decreased by the presence of ACC deaminase, either by applying P. migulae 8R6 to the soil or via acdS expression in the transgenic lines, at both moderate and high salt concentrations (EC 15 and 20 dS m−1). The line expressing acdS under the control of the CaMV 35S promoter had the lowest levels of ethylene at the highest salt concentration.
The effect of ACC deaminase on ethylene production in C. sativa under salt stress (NaCl; 15 and 20 dS m−1). Salt was applied 20 days after sowing and plant material collected 13 days afterward. Soil was treated with buffer (control) or the P. migulae 8R6 strain producing ACC deaminase. Transgenic lines tested were independent, single insert, homozygous lines expressing the Pseudomonas sp. UW4 acdS gene under the control of either the rolD or the CaMV 35S promoter. Error bars indicate standard error (n = 3 biological replicates). Significant differences between groups were detected using a two-way ANOVA and Tukey post-test. Values that are significantly different (p < 0.05) are indicated by letters. Plots were drawn using SigmaPlot ver. 13.0 (Systat Software, Inc., San Jose, USA, www.systatsoftware.com).
As expected, shoot length, fresh and dry weight decreased as the salt concentration increased regardless of the treatment; however, the presence of ACC deaminase delivered either by PGPB or via acdS expression in transgenic lines significantly reduced the impact of salt stress (Fig. 2). The reduction in shoot length was significantly less for plants grown in soil treated with P. migulae 8R6 or P. fluorescens YsS6, but not with the corresponding acdS-mutants, as was also the case for transgenic lines expressing acdS under the control of the rolD promoter. Shoot fresh and dry weight responded similarly and the beneficial effect was also observed in lines expressing acdS under the control of the CaMV 35S promoter. In addition, the control plants were severely wilted 20 days after treatment with the higher salt concentration (EC 20 dS m−1), whereas plants expressing acdS or treated with wild-type PGPB were not18.
The effect of ACC deaminase on C. sativa shoot length (A), fresh weight (B), and dry weight (C) in the absence or presence of salt stress (NaCl; 15 and 20 dS m−1). Soil was treated with buffer (control), P. migulae 8R6 producing ACC deaminase or a derived acdS mutant 8R6 M, P. fluorescens YsS6 producing ACC deaminase or a derived acdS mutant YsS6 M. Transgenic lines tested were independent, single insert, homozygous lines expressing the Pseudomonas sp. UW4 acdS gene under the control of either the rolD or the CaMV 35S promoter. Salt was applied 20 days after sowing and shoots were harvested 20 days afterward. Error bars indicate standard error (n = 10 biological replicates). Significant differences between groups were detected using a two-way ANOVA and Tukey post-test. Values that are significantly different (p < 0.05) from the control are indicated by asterisks (*) above the bars. Plots were drawn using SigmaPlot ver. 13.0 (Systat Software, Inc., San Jose, USA, www.systatsoftware.com).
Chlorophyll content and photosynthetic yield (Fv/Fm) illustrate the level of activity in the photosystem reaction centers. Chlorophyll content was significantly higher in the acdS transgenic lines and in plants grown in soil treated with the wild-type PGPB under salt stress, but not with the acdS-mutants (Fig. 3). Photosynthetic yield was maintained to an even higher degree and was especially apparent at the higher salt concentration (EC 20 dS m−1). The effect was observed with all treatments, including the PGPB acdS-mutants as these bacteria possess additional mechanisms to promote plant growth and health15,19.
The effect of ACC deaminase on C. sativa photosynthetic capacity in the absence or presence of salt stress (NaCl; 15 and 20 dS m−1). Panels show chlorophyll content and photosynthetic yield 33 days after sowing with salt being applied 20 days after sowing. Soil was treated with buffer (control), P. migulae 8R6 producing ACC deaminase or a derived acdS mutant 8R6 M, P. fluorescens YsS6 producing ACC deaminase or a derived acdS mutant YsS6 M. Transgenic lines tested were independent, single insert, homozygous lines expressing the Pseudomonas sp. UW4 acdS gene under the control of either the rolD or the CaMV 35S promoter. Error bars indicate standard error (n = 10 biological replicates). Significant differences between groups were detected using a two-way ANOVA and Tukey post-test. Values that are significantly different (p < 0.05) from the control are indicated by asterisks (*) above the bars. Plots were drawn using SigmaPlot ver. 13.0 (Systat Software, Inc., San Jose, USA, www.systatsoftware.com).
ACC deaminase had little effect on the concentration of macro- and micro-elements in vegetative tissues. The levels of some macro-elements changed with increasing salinity; for example, the sodium levels rose sharply and the potassium and calcium levels declined; however, expression of acdS or treatment of soil with P. migulae 8R6 had little or no impact on the levels or trends (Fig. 4). With respect to the micro-elements, the boron concentration was significantly elevated by acdS expression and the presence of P. migulae 8R6, but only in the absence of salt.
The effect of ACC deaminase on shoot element content in C. sativa grown in the absence or presence of salt. Soil was treated with buffer (control) or P. migulae 8R6 producing ACC deaminase. Transgenic lines tested were independent, single insert, homozygous lines expressing the Pseudomonas sp. UW4 acdS gene under the control of either the rolD or the CaMV 35S promoter. Panels show macro- and micro-element content in shoots of plants 33 days after sowing that were irrigated with either tap water or with a salt solution (15 or 20 dS m−1); application of the salt solution began 20 days after sowing. Significant differences between groups were detected using a two-way ANOVA and Tukey post-test. Values that are significantly different (p < 0.05) from the control are indicated by asterisks (*). Error bars indicate standard error (n = 3 biological replicates). Plots were drawn using SigmaPlot ver. 13.0 (Systat Software, Inc., San Jose, USA, www.systatsoftware.com).
Global changes in C. sativa transcription in response to salt treatment
ACC deaminase was demonstrated to improve the tolerance of C. sativa to salinity stress. To further explore this phenomenon, the transcriptome of the above ground (vegetative) tissue of the wild-type DH55 line was compared to tissues from lines expressing acdS under the control of the root-specific promoter (rolD) or the constitutive promoter (CaMV 35S), as well as to tissues from plants grown in soil treated with P. migulae 8R6. The rolD promoter was not entirely root-specific, since acdS expression in vegetative tissues was approximately one-half the level observed in roots. However, in vegetative tissues, the expression of the acdS gene was 60 times higher under the control of the CaMV 35S promoter than under the rolD promoter (data not shown).
The gene expression patterns were defined in terms of the impact ACC deaminase had on the expression normally observed in the non-trangenic (wild-type) or untreated (not grown in soil amended with PGPB) control line under salt stress. These patterns include: “induced” or “down-regulated” when the expression of the gene was not affected by salt stress in the control, but was altered in the transgenic or PGPB-treated lines; “less induced” when the gene was induced in the control, but to a lesser degree in the transgenic or PGPB-treated lines and similarly for “less down-regulated”; “more induced” when the gene was induced in the control, but to a greater degree in the transgenic or PGPB-treated lines and similarly for “more down-regulated”.
In vegetative tissues of plants treated with P. migulae 8R6, 29 genes were induced and 81 genes were down-regulated, while 226 genes were less induced and 18 genes were less down-regulated compared to the control (DH55 not treated with P. migulae 8R6) in the presence of salt (FDR ≤ 0.05 and expression ≥ twofold; Fig. 5; Supplemental Data 2). In the 35S::acdS line, 27 genes were induced, 170 genes were down-regulated, 8 genes were more induced, 89 genes were less induced, 7 genes were more down-regulated and 10 genes were less down-regulated. In the rolD::acdS line, 42 genes were induced, 368 genes were down-regulated, 12 genes were more induced, 357 genes were less induced, 26 genes were more down-regulated and 3 genes were less down-regulated compared to the control in the presence of salt. It is interesting to note that expression of acdS under the control of the rolD promoter had a much greater effect on the expression of salt-responsive genes than expression under the control of the much stronger CaMV 35S promoter. In total, about 300 genes were differentially-regulated in the 35S::acdS line, while the expression of about 800 genes was affected in the rolD::acdS line. Approximately, three times as many genes were less induced or more down-regulated in the rolD::acdS line compared to the 35S::acdS line or plants grown in soil treated with P. migulae 8R6 after exposure to salt.
Differential-expression genes in vegetative tissue from C. sativa lines expressing acdS under the control of the rolD promoter or the constitutive CaMV 35S promoter or from plants grown in soil treated with P. migulae 8R6 in the presence of salt (NaCl, 15 dS m−1). Left hand panel: Pie diagrams providing the number of genes that were differentially-regulated compared to the non-transgenic, untreated control. Right hand panel: plots representing the number of genes that were differentially-expressed on each of the C. sativa sub-genomes (top) and genome partitioning defined as the proportion of times when homeologous genes on one, two or three genomes were differentially-expressed (bottom). Genes with an FDR and p value ≤ 0.05 and with an absolute value of log 2 CPM higher than 1 were considered significant. Plots were drawn using SigmaPlot ver. 13.0 (Systat Software, Inc., San Jose, USA, www.systatsoftware.com).
Of the differentially-regulated genes in plants treated with P. migulae 8R6, approximately one third were assigned to each sub-genome with 121 assigned to Genome I, 110 to Genome II and 121 to Genome III. The fractional distribution in the rolD::acdS line was similar with 283 genes assigned to Genome I, 260 to Genome II and 284 to Genome III (Fig. 5). However in the 35S::acdS line, 120 genes were assigned to Genome III with only 101 assigned to Genome I and 89 to Genome II. An indication of partial genome partitioning in response to salt stress was found by examining the expression of homeologous genes. In plants grown in soil treated with P. migulae 8R6, 47% of the differentially-expressed genes were assigned to only one sub-genome, while 26% of the differentially-expressed genes (84 genes—42 homeologous pairs) were assigned to two sub-genomes, and 27% (87 genes—29 homeologous groups) were assigned to all three sub-genomes. In the 35S::acdS line, 60% were assigned to only one sub-genome, while 30% (76 genes—38 homeologous pairs) were assigned to two sub-genomes, and only 10% (24 genes—8 homeologous groups) were assigned to all three sub-genomes. In the rolD::acdS line, 33% (212 genes—106 homeologous pairs) were assigned to two sub-genomes, whereas 21% (132 genes—44 homeologous groups) were assigned to all three sub-genomes with the remainder being assigned to only one sub-genome (46%) (Fig. 5).
To confirm the RNA-Seq data, a set of differentially-expressed genes was chosen for quantitative ddPCR analysis (Supplemental Information 3). The expression profiles of the genes were very similar regardless of the method. A complete annotation of the genes that were differentially-expressed in vegetative tissues of these lines can be found in Supplemental Data 4 and are discussed further below. Heat maps superimposed on MapMan GO pathways were used to visualize systems that were altered in response to salt stress and are available in Supplemental Information 4.
Systems and biochemical pathways altered by ACC deaminase during response to salt
Photosynthesis
Salt treatment negatively impacts photosynthesis and the expression of genes related to photosynthesis; however, ACC deaminase positively affected the expression of some genes involved in photosynthesis upon salt treatment (Supplemental Data 2). Csa08g050560 (photosystem I subunit D-1) was less down-regulated and three genes involved in the photosystem II light harvesting complex were less down-regulated (Csa14g037480 and Csa06g036870) or induced (Csa01g001010) in vegetative tissues of plants grown in soil treated with P. migulae 8R6 compared to the control DH55 line under salt stress. In the 35S::acdS line, Csa17g025490 (chlorophyllase 1) was down-regulated compared to the wild-type DH55 line during salt stress.
Cell wall biosynthesis/modification and secondary metabolism
The plant cell wall provides a structural framework for the mechanical and physical properties of the cell and determines the amount of water uptake and cell expansion available to support plant growth30. In the rolD::acdS line, which expressed acdS at low levels in vegetative tissues, cell wall-related genes were mostly down-regulated (40 genes down-regulated, 8 genes less induced, 10 genes more down-regulated and only 2 genes induced) in response to salt stress compared to wild-type DH55 (Supplemental Data 2). Interestingly, in the 35S::acdS line, which has 60-fold more acdS transcripts in vegetative tissues than the rolD::acdS line, only 9 cell wall-related genes were differentially-regulated (3 genes down-regulated, 2 genes more down-regulated, 1 gene less down-regulated and 3 genes induced).
Secondary metabolites accumulate during salt stress, for example anthocyanins and flavonoids are generated in response to salinity stress and during leaf senescence in A. thaliana31. However, genes involved in the synthesis of secondary metabolites in plants grown in soil treated with P. migulae 8R6, or in the lines expressing acdS were generally less-induced or down-regulated (Supplemental Data 2).
Phytohormones
Hormones are essential for plant growth and development, and are essential for adaptation to adverse biotic and abiotic environmental factors. ACC deaminase directly impacts the production of ethylene by limiting the amount of its immediate precursor. Interestingly, this was also found to indirectly affect the expression of genes involved in the synthesis, regulation or responses associated with other plant hormones as described below.
Auxin
Five genes related to auxin metabolism, including those encoding IAA-amino acid hydrolase ILR1-like 4 (Csa14g060060) and IAA-amino acid conjugate hydrolase ILR1-like 6 (Csa14g049310, Csa03g046940 and Csa17g070920) were less induced after salt treatment in the vegetative tissues of plants grown in soil treated with P. migulae 8R6. Five auxin-responsive genes encoding members of the Small Auxin Up RNA (SAUR) family, including Csa07g039520, Csa09g076010 and Csa16g035110 (SAUR 78), were induced or less down-regulated in the 35S::acdS line, while Csa17g042390 (SAUR 66) was induced in the rolD::acdS line. In the rolD::acdS line, five auxin-responsive genes encoding GH3 family proteins were down-regulated (Csa16g044890, Csa09g087450, Csa07g053230, Csa08g005200) or less induced (Csa10g003770) (Supplemental Data 2).
Gibberellic acid (GA)
Csa01g002510 and Csa07g046090, which encode GA-Stimulated Arabidopsis (GASA) 5 and 6, respectively, were the only GA-dependent genes that were differentially-expressed. These were less down-regulated and only in plants grown in soil treated with P. migulae 8R6 (Supplemental Data 2).
Cytokinin
Cytokinin oxidase 3 (CKX3; Csa11g090460) catalyzes the degradation of cytokinins and was down-regulated fivefold in the 35S::acdS line. Csa11g090460 and one of its homeologues (Csa02g061320) were less induced or down-regulated (78-fold), respectively, in the rolD::acdS under salt stress compared to the wild-type DH55. Csa02g061320 was up-regulated 20-fold in the non-transgenic line by salt stress, again representing significant attenuation of the stress response (Supplemental Data 2).
Abscisic acid (ABA)
Genes encoding positive regulators of ABA signalling, namely 9-cis-epoxycarotenoid dioxygenase 3 (NCED; Csa19g021150 and Csa15g018820), which is involved in ABA biosynthesis, and ABA Insensitive 2 (Csa02g061490) were less induced, while that encoding ABA-induced PP2C5 (Csa05g012640) was down-regulated upon salt stress in plants grown in soil treated with P. migulae 8R6. The expression of genes encoding negative regulators of ABA signal transduction, including Highly ABA-Induced PP2C protein 1 (three homeologous genes Csa02g064780, Csa18g032370 and Csa11g092890), Highly ABA-Induced PP2C protein 2 (three homeologous genes Csa03g010830, Csa17g010800 and Csa14g008830) and PP2CA (Csa15g015150) were still induced by salt stress, but approximately threefold less in these plants. These genes were induced up to 250-fold in the control line in response to salt stress in the absence of PGPB (Supplemental Data 2).
Contrary to the treatment with P. migulae 8R6, expression of acdS in C. sativa did not greatly affect the expression of genes involved in ABA biosynthesis or signal transduction. Csa04g054460, another homeologue of Csa05g012640 encoding PP2C5, a positive regulator of ABA signal transduction, was less induced (fivefold) compared to the non-transgenic line, but only in the rolD::acdS line. Csa14g008830 encodes ABA-induced PP2C, a negative regulator of ABA signaling, and was less induced both in plants grown in soil treated with P. migulae 8R6 and in the acdS transgenic lines. This gene was induced 70-fold in the wild-type DH55 control by salt stress (Supplemental Data 2).
Ethylene
As shown above, ACC deaminase reduces ethylene production in plants exposed to salt stress. In accordance, genes encoding Ethylene Responsive Factors (ERF), such as Csa03g031680 and Csa17g039230 encoding ERF11, Csa01g017750 (ERF4), Csa14g063540 and Csa17g093440 encoding ERF8, as well as Csa03g046520 and Csa17g069360 (RAP2.6), which are involved in ethylene-activated signaling, were less induced in vegetative tissue of plants grown in soil treated with P. migulae 8R6 compared to the untreated control during salt stress. In the 35S::acdS line, two other homeologues of Csa01g017750 encoding ERF4 (Csa15g019690 and Csa19g021900), Csa20g081680 (ERF2), as well as Csa11g018190 and Csa10g016710 (ACC synthase 7) were down-regulated or less induced after salt treatment. Similarly, Csa19g033660 (ERF1), Csa03g031680 (ERF11), Csa12g047140 and Csa11g064550 (ethylene responsive element binding factor 1; EBF1), as well as Csa20g081680, Csa18g002590 and Csa14g006260 (EBF2), which are involved in ethylene signaling, were down-regulated or less induced in the rolD::acdS line after salt treatment. Many genes involved in ethylene biosynthesis, including Csa14g006260 and Csa17g008270 (ACC oxidase 4), Csa03g001750 and Csa17g001950 (ACC synthase 2), as well as Csa11g018190, Csa12g025860 and Csa10g016710 (ACC synthase 7) were also down-regulated or less induced; these were induced up to 500-fold in wild-type DH55 after salt treatment. In fact, Csa08g022870 (Ethylene and Salt Inducible 3) was the only ethylene-related gene that was induced and only in the 35S::acdS line (Supplemental Data 2).
Jasmonic acid (JA)
Several genes involved in JA biosynthesis were down-regulated in plants grown in soil treated with P. migulae 8R6 compared to the untreated control plants after salt treatment, while no genes related to JA biosynthesis or signaling were differentially-expressed in the vegetative tissues of the acdS transgenic lines. Genes involved in JA biosynthesis that were down-regulated or less induced in the P. migulae 8R6-treated plants included Csa17g023140 and Csa03g021200 (lipoxygenase 3), Csa07g039620 (lipoxygenase 4), Csa19g040810 and Csa01g038310 (oxophytodienoate-reductase 3), as well as Csa06g005320, Csa09g008880 and Csa03g038080 (allene oxide cyclase 1, 2 and 3). Most of these genes were highly induced (up to 26-fold) by salt stress in the untreated control (Supplemental Data 2).
Protein degradation and cellular turnover
The environmental conditions surrounding plants are constantly changing and post-translational regulation of protein levels (regulatory and functional) is an important mechanism underlying adaptation32. The ubiquitination pathway in particular plays a role in altering the plant proteome in response to environmental stresses33,34 and many genes involved in protein degradation/turnover were less induced, both in plants treated with P. migulae 8R6 and in the acdS transgenic lines, compared to the untreated, non-transgenic control during salt stress (Supplemental Data 2) providing evidence for a common mechanism regardless of the means for ACC deaminase delivery. Fully, 25% of the genes commonly down-regulated in the acdS transgenic lines were associated with protein degradation.
Stress response
Abiotic stress
Plant stress responses and development are tightly connected processes and their precise regulation is required to maintain fitness when a stress is encountered. Eight genes related to different abiotic stresses were down-regulated or less induced in vegetative tissues of plants grown in soil treated with P. migulae 8R6 during salt stress, including two (Csa18g023200 and Csa02g045340) encoding Responsive to Desiccation 29B (RD29B). Furthermore, three genes encoding Responsive to ABA 18 (RAB18; Csa18g042170, Csa11g104590 and Csa02g076390) were also less induced in these plants (Supplemental Data 2). The expression of both RAB18 and RD29B was highly induced 68 and 244-fold, respectively, in vegetative tissues of untreated C. sativa upon exposure to salt stress (Supplemental Data 2).
Although Late Embryogenesis Abundant (LEA) proteins are classified under the GO category “development process”, many members of this family play an important role in the abiotic stress response and stress tolerance35. Seven genes encoding LEA proteins were less induced and two were down-regulated in plants grown in soil treated with P. migulae 8R6 compared to the control (Supplemental Data 2). These were highly up-regulated (up to 185-fold) by salt stress in untreated C. sativa suggesting that ACC deaminase has impacted overall stress levels. In the rolD::acdS line, three homologous genes encoding LEA5 were less induced (Csa13g054850 and Csa02g005610) or down-regulated (Csa08g052090), as was a gene encoding NDR1/HIN1-like 6 (NHL6; Csa07g029570, less induced) which is also responsive to abiotic stress36.
Biotic stress
Genes encoding AvrRrt2-Induced Gene1 (Csa03g038080) and Receptor-like protein 22 (Csa16g008730), which are involved in the response to bacterial pathogens, were the only genes related to biotic stress that were differentially-regulated in plants grown in soil treated with P. migulae 8R6. However, 11 genes related to biotic stress were less induced or down-regulated in the 35S::acdS line under salt stress, including Csa16g038950 (Pathogenesis-Related Protein; PR5), Csa11g001970 and Csa10g001780 (Enhanced Disease Susceptibility 5; EDS5) and Csa11g032780 (Suppressor of npr1, constitutive 1) which were all less induced compared to the non-transgenic line after salt stress. In the rolD::acdS line, 28 genes related to biotic stress were less induced or down-regulated, including those encoding PR1 (Csa01g035420, Csa19g044900 and Csa15g064830), PR5 (Csa16g038950, Csa07g046470 and Csa09g079640), a PR5-like protein (Csa09g079620), and EDS5 (Csa10g001780 and Csa11g001970) (Supplemental Data 2).
Signalling
Transcription factors (TF)
The expression of several genes encoding NAC (NAM, ATAF1/2, CUC2-domain) transcription factors were less induced or down-regulated in plants grown in soil treated with P. migulae 8R6 and in the acdS transgenic lines compared to the wild-type control during salt treatment. For example, three homoleogous genes (Csa12g023460, Csa11g016750 and Csa10g015440) which encode Responsive to Desiccation 26 (RD26, NAC domain-containing 72) were induced threefold less in P. migulae 8R6-treated plants. Genes encoding NAC019 (Csa17g093080, Csa14g063210, and Csa03g060000), NAC055 (Csa01g018090 and Csa19g022270) and NAC102 (Csa18g037380) were less induced in P. migulae 8R6-treated plants, as well as in the rolD::acdS (NAC019 and NAC055) and 35S::acdS lines (NAC102). However, all three homeologues encoding NAC019 were less induced in the P. migulae 8R6-treated plants, while only the homeologue on sub-genome III (Csa03g060000) was less induced in the rolD::acdS line. Genes encoding NAC102 (Csa18g037380), NAC067 (Csa02g004650) and NAC043 (Csa05g002390), were less induced or down-regulated in the 35S::acdS line. In the rolD::acdS line, genes encoding NAC019 (Csa03g060000) and NAC055 (Csa19g022270), and other NAC proteins such as NAC036 (Csa19g053810), NAC042 (Csa05g007460), NAC067 (Csa02g004650), NAC069 (Csa02g004670) and NAC087 (Csa13g021170) were less induced or down-regulated (Supplemental Data 2).
In total, 48 genes encoding other TFs, mostly from the AP2/EREBP, WRKY, MYB and zinc finger (ZF) families, including ERF17 (Csa03g023030), Dehydration Responsive Element-binding Protein 2A (DREB2A; Csa13g007380, Csa20g006670 and Csa08g059070), and AP2.6 (Csa03g046520 and Csa17g069360) were less induced or down-regulated in plants grown in soil treated with P. migulae 8R6. All of these genes were highly up-regulated (up to 116-fold) by salt stress in untreated control plants in response to salt. Two homeologous genes (Csa18g029750 and Csa11g090310) and Csa18g024230 encoding a MYB-like TF were the only TF genes that were induced in plants grown in soil treated with P. migulae 8R6 (Supplemental Data 2). Of the genes encoding MYB TFs that were less induced in the P. migulae 8R6-treated plants under salt stress, some of the more notable ones encoded MYB20 (Csa16g023660), MYB74 (Csa02g009840, Csa08g049290 and Csa13g053050), MYB102 (Csa11g025000, Csa12g036900 and Csa10g021970) and MYB108 (Csa01g007530). In response to salt stress in untreated C. sativa, genes encoding MYB20 and MYB108 were up-regulated tenfold, MYB102 up to 40-fold and AtMYB74 up to 1500-fold (Supplemental Data 2).
In total, 16 and 64 genes encoding TFs (other than NAC) were less induced or down-regulated in the 35S::acdS and rolD::acdS transgenic lines, respectively, with most encoding members of the AP2/EREBP, WRKY, MYB, ZF and basic Helix-Loop-Helix (bHLH) families. Among those in common, four genes were less induced, including Csa18g010220 (bHLH 92) which is involved in regulating the circadian clock in A. thaliana37, or down-regulated, such as Csa02g073920 which encodes WRKY51 and is involved in repression of JA signaling38. Over-expression of some genes encoding WRKY TFs increases tolerance against some biotic and abiotic stresses; however, the beneficial effects were often accompanied by some undesirable phenotypes39. In the rolD::acdS line, genes encoding WRKY25, 30, 31, 33, 38, 47, 50, 51, 53 and 70 were less induced or down-regulated (2 to 40-fold) compared to the wild-type line under salt stress (Supplemental Data 2).
Csa19g025930 encodes a TF with a TCP domain (Branched 1) and was less induced in the 35S::acdS line. No genes encoding AP2/EREBP TFs were differentially-regulated in the 35S::acdS line, while two genes encoding bZIP TFs (Csa06g033960 and Csa09g070700) were less down-regulated and one encoding a Homeobox protein (Csa11g094320) were down-regulated. Eleven genes encoding AP2/EREBP TFs were less induced or down-regulated in the rolD::acdS line (Supplemental Data 2); these included Csa14g016230 (Regulator of the ATPase of the vascular membrane, RAV1), Csa16g028940 and Csa07g034530 (RAV2 or Tempranillo 2), Csa14g032810 and Csa03g029510 (Tempranillo 1) which were down-regulated and Csa12g027700 (C-Repeat/DRE binding factor 2; CBF2) which was induced fourfold less compared to the non-transgenic line after salt treatment. Four genes encoding B3 or bHLH TFs were induced or less down regulated.
Discussion
In a previous study, C. sativa plants grown in soil amended with PGPB that produce ACC deaminase or lines expressing the bacterial acdS gene grew better in a salt mixture formulated to mimic naturally saline soils on the North American prairies16. Most other published studies on salinity tolerance use only NaCl, therefore, the studies described herein and a prior study on the root transcriptome18 used NaCl at the same EC values as the natural saline compositions. Regardless of the type of salt treatment, C. sativa lines expressing acdS or plants grown in soil treated with PGPB exhibited less decline in shoot length and weight, and reduced impact on chlorophyll content and photosynthetic yield. PGPB mutants lacking acdS also positively impacted certain aspects related to salinity tolerance, such as photosynthetic yield, though to a lesser degree than the wild-type strains, which may be attributed to the ability of these strains to promote plant health by producing plant growth-regulators and increasing the solubility of soil minerals40. It is noted here that transcript abundance generally, but does not always, correlate with protein levels. However, this type of study permits examination of a broad suite of pathways and provides clues as to the mechanisms underlying tolerance to salinity stress as discussed below.
Impact of ACC deaminase on photosynthesis
Salt treatment negatively impacts photosynthesis and the expression of genes related to photosynthesis. ACC deaminase reduced the negative effect of salt on photosynthesis by inducing the expression of genes involved in photosystem I or II and the light harvesting complex, as in plants grown in soil treated with P. migulae 8R6, or by reducing the expression of chlorophyllase, as in the 35S::acdS line. Chlorophyllase is involved in chlorophyll degradation41 and reducing its activity improves photosynthetic efficiency and plant productivity under stress conditions42. While photosynthetic capacity improved in the rolD::acdS line, no significant differences were observed in the expression of photosynthetic genes in the vegetative tissues of this line after salt treatment compared to the non-transgenic line. Interestingly, photosynthetic genes are also expressed in root tissues12,43 and ACC deaminase delivered by PGPB or in transgenic C. sativa lines attenuates the negative impact of salt on their expression18.
This raises the possibility that sensing of salinity stress in roots and subsequent alteration in the expression of photosynthesis-related genes in roots affects photosynthetic capacity in vegetative tissues. The root meristem harbors cells that sense salt stress44and this can direct the behavior of other tissues, such as the closure of leaf stomata within seconds after exposure of roots to salt45.
Phytohormones
Ethylene signaling is essential for regulating plant growth and development in response to different environmental stresses and, specifically, is important for acclimation to saline conditions46,47. However, elevated levels of ethylene impede plant growth leading to smaller rosettes, earlier flowering and reduced seed production48, as well as the onset of senescence, chlorosis and leaf abscission14, and a negative feedback mechanism by ethylene has been proposed in salinity stress49. In the current study, the level of ethylene was found to be lower in C. sativa plants exposed to salt which seemed incongruent with its role in salinity stress; however, it should be noted that we sampled 13 days after the application of salt and very few studies have examined the response to chronic salt exposure50. Furthermore, increases in ethylene production associated with salt exposure recover to control levels within 24 h after treatment in salt-tolerant species51. Camelina sativa is comparatively salt-tolerant and this data supports the notion that ethylene production/sensitivity may already be highly regulated in this species, but that ACC deaminase helps to reduce levels under salt stress even further. Indeed, the expression of many genes involved in ethylene synthesis or ethylene signaling are highly induced in C. sativa by salt treatment, for example ACC synthase 2 and 7 were induced almost 400- and 500-fold, respectively12. The negative effect of ACC synthase and ACC oxidase, and resultant ethylene biosynthesis on salt tolerance has been documented49,52. The effect of high levels of stress ethylene can be reduced by ACC deaminase application which restores normal plant growth habit while inducing systemic tolerance under stress14,15,16,53. In the current study, not only ethylene, but the expression of genes involved in ethylene synthesis or ethylene signaling was reduced by ACC deaminase application. This indicated that the ability of ACC deaminase to diminish or attenuate the ethylene response goes beyond simply lowering ethylene levels, but also impacts the expression of genes involved in its synthesis and responsiveness.
Comparison of the phenotypes and the expressed genes in the 35S::acdS and rolD::acdS lines indicated that moderate acdS expression was more effective than high expression in eliminating the negative effects of ethylene production caused by salt stress. The expression of acdS was 60-fold higher in the 35S::acdS line compared to the rolD::acdS line; however, the number of ethylene-related genes that were differentially-expressed (mostly down-regulated or less induced) was three times higher in the rolD::acdS line. In addition, the down-regulation or the attenuation of induction was more severe in the rolD::acdS line, for example the three homeologous genes encoding ACC synthase 7- were 27-fold less induced in the rolD::acdS line, but only five-fold less in the 35S::acdS line compared to the wild-type line upon salt treatment. Some controversy exists with respect to the role of ethylene in salt stress tolerance in different plants, for example over-expression of genes encoding a number of ERFs enhanced salt tolerance in A. thaliana54. In the transgenic lines expressing acdS, genes involved in ethylene biosynthesis, such as those encoding ACC synthase or ACC oxidase, were less induced compared to the wild-type line after salt treatment. However, in plants grown in soil treated with P. migulae 8R6, most of the genes related to ethylene that were less induced encoded negative regulators of ethylene signaling55. This seems counter-intuitive as down-regulation of a negative regulator should enhance the effect of ethylene. However, several of these regulators are also involved in crosstalk between ethylene and ABA signaling, such as ERF4, ERF7 and ERF1255,56, and, therefore, the actual mechanism and interplay is likely far more complex.
Auxin signalling and cross-talk also appear to have been affected by ACC deaminase as several auxin-responsive genes encoding members of the Small Auxin Up RNA (SAUR) family were up-regulated in the 35S::acdS and rolD::acdS lines. SAUR 76–78 affect ethylene receptor signaling and promote plant growth57. Furthermore, in the rolD::acdS line and in plants grown in soil treated with P. migulae 8R6, several auxin-responsive genes encoding GH3 family proteins and/or IAA-amino acid hydrolases, which regulate auxin homeostasis by conjugating excess auxins to amino acids58,59, were less induced. Auxin conjugates compartmentalize auxins to sequester excess IAA or to protect the free acid from peroxidative degradation58,60.
The expression of ABA-dependent genes is highly up-regulated (up to 230-fold) in wild type C. sativa in response to salt treatment12. However, in vegetative tissues of plants grown in soil treated with P. migulae 8R6, genes encoding enzymes involved in ABA biosynthesis (NCED) or positive regulators of ABA signal transduction (Insensitive 2 and PP2C5) were less induced, as were several genes encoding negative regulators of ABA signal transduction. This implies that ABA is important for salt tolerance, but that a balance between the positive effect of ABA on stress tolerance and its negative effect on growth and development is necessary. After sensing salinity, two type of reactions (immediate and long-term response) are coordinated by ABA. The immediate response involves inhibition of cell elongation; however, upon prolonged exposure to salt stress, ABA promotes plant growth, but at a lower rate than in the absence of salt stress44. The extent of growth recovery depends on the capacity of the plant for acclimation44,61. Contrary to the treatment with P. migulae 8R6, expression of acdS in transgenic C. sativa did not greatly affect the expression of genes involved in ABA biosynthesis or signal transduction indicating that ACC deaminase may not be the only reason for the PGPB-mediated effect.
In general, treatment of soil with a P. migulae 8R6 strain producing ACC deaminase negatively affected ethylene signaling, auxin and JA biosynthesis and signalling. In addition, ABA production and signalling was also altered in a way that likely reduces the adverse effect of ABA on plant growth and productivity during salt stress. However, PGPB treatment seems to have had a positive effect on the regulation of the GA signaling. In plants expressing acdS, the expression of the genes involved in auxin signalling was positively affected, while the expression of genes involved in cytokinin degradation and ethylene biosynthesis was negatively affected. Fine-tuning of ABA signaling may also be occurring.
Stress response and leaf senescence
Exposure to biotic and/or abiotic stresses redirects plant physiological activity from vegetative growth towards resistance or defense activation. Tuning the regulation of the corresponding genes is crucial to maximizing plant fitness while not unnecessarily compromising plant growth and development62,63. For example, accumulation of plant secondary metabolites, such as anthocyanins and flavonoids, which increase salinity resistance in plants, accompanies leaf senescence and reduces plant yield and biomass31,64. The overall trend in the transcriptome data showed a reduction in ethylene production and down-regulation of genes involved in the accumulation of stress-related secondary metabolites in the presence of ACC deaminase.
In addition to the impact on secondary metabolism, several genes encoding NAC transcription factors, including RD26, NAC019, NAC055 and NAC102, and genes encoding RD29B and Responsive to ABA 18, were all down-regulated or less induced in plants grown in soil treated with P. migulae 8R6 and the transgenic lines during salt stress. These transcription factors regulate the biotic and abiotic stress response and play essential roles in senescence and chlorophyll degradation65,66. Salinity and ethylene both induce leaf senescence67. rd29a and rd29b mutant lines display greater root growth, photosynthesis, and water use efficiency under salt stress68 and RD26 is a positive regulator of age and dark-induced leaf senescence in addition to its role in desiccation69. NAC019 functions downstream of ethylene insensitive 2 (EIN2) and EIN3, ethylene-activated transcription factors necessary for the enhanced salt tolerance70; however, NAC019, NAC055 and NAC102 are positive regulators of leaf senescence66,67,71. Two genes encoding Yellow-leaf Specific Gene 5 and 9, and a gene encoding ULTRAPETALA1 were also less induced in plants grown in soil treated with P. miguale 8R6. The expression of these genes was up-regulated up to 244-fold by salt in vegetative tissues of untreated C. sativa. Yellow-leaf Specific Gene 5 and 9 are involved in leaf senescence and ULTRAPETALA1 acts as a negative regulator of shoot, floral meristem size and flower development72.
In addition to the lower induction or down-regulation of genes encoding NAC TFs in the rolD::acdS line, genes encoding RAV1 (Related to ABI3/VP11), Tempranillo 1 and Tempranillo 2, and several WRKY TFs (WRKY25, 30, 31, 33, 38, 47, 50, 51, 53 and 70) were less induced or down-regulated. The expression of these genes was up-regulated up to 1000-fold by salt stress in wild-type C. sativa. RAV1 is a negative regulator of growth and a positive regulator of leaf senescence. A. thaliana lines over-expressing RAV1 exhibit strong growth retardation and a semi-dwarf stature73. TEM1 and TEM2 delay flowering and induce leaf senescence when over-expressed in A. thaliana and seed germination is more inhibited by salt stress73,74. Over-expression of some WRKY genes was reported to increase tolerance to some biotic and abiotic stresses; however, the beneficial effects were often accompanied by unwanted phenotypes39. WRKY TFs are the second largest family of TFs involved in senescence75 with WRKY22, WRKY30, WRKY53, WRKY54, WRKY70, and WRKY75 playing important roles in leaf senescence76,77,78,79. Genes encoding ZAT7 and ZF protein 1, which are negative regulators of growth under abiotic stress conditions80, were also down-regulated or less induced in the rolD::acdS line under salt stress.
Conclusions
The application of ACC deaminase by P. migulae 8R6 or in acdS transgenic lines caused a reduction in the expression of stress-responsive genes that are highly induced by salt to levels that may lower or indicate a reduced negative effect on the plant. This is in agreement with earlier observations that the impact of salinity stress on growth and productivity was reduced by treatment of soil with P. migulae 8R6 or in acdS transgenic lines16. In many cases, stress adaptation comes at the expense of growth and productivity, therefore, it is necessary for plants to develop resilient systems to optimize the trade-off between short term survival and long-term growth. The physiological and transcriptome data clearly demonstrated that decreasing the concentration of stress ethylene through the application of ACC deaminase can significantly increase the extent of plant growth and productivity under salt stress in C. sativa. Expressing the acdS gene under the control of different promoters or through the application of endophytic PGPB sometimes affected the expression of different homeologous genes indicating that in polyploid plants studying the effect of stress on gene expression in different sub-genomes is important. This information may be used as a resource for breeders to target the correct homeologue in C. sativa to obtain the desired effect on salinity stress. However, approaches, such as the use of ACC deaminase, that regulate a network of genes may be more effective than targeting a single gene to provide stress tolerance and reduce the negative impact on growth and yield production.
Data availability
The RNA-Seq data were deposited to NCBI Gene Expression Omnibus under the accession code GSE132600.
References
Blackshaw, R. et al. Alternative oilseed crops for biodiesel feedstock on the Canadian prairies. Can. J. Plant Sci. 91, 889–896. https://doi.org/10.4141/cjps2011-002 (2011).
Li, X. & Mupondwa, E. Life cycle assessment of camelina oil derived biodiesel and jet fuel in the Canadian prairies. Sci. Total Environ. 481, 17–26. https://doi.org/10.1016/j.scitotenv.2014.02.003 (2014).
Hutcheon, C. et al. Polyploid genome of Camelina sativa revealed by isolation of fatty acid synthesis genes. BMC Plant Biol. 10, 233–247. https://doi.org/10.1186/1471-2229-10-233 (2010).
Kagale, S. et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 5, e3706. https://doi.org/10.1038/ncomms4706 (2014).
Kanth, B. K., Kumari, S., Choi, S. H., Ha, H. J. & Lee, G. J. Generation and analysis of expressed sequence tags (ESTs) of Camelina sativa to mine drought stress-responsive genes. Biochem. Biophys. Res. Commun. 467(1), 83–93. https://doi.org/10.1016/j.bbrc.2015.09.116 (2015).
Poudel, S., Aryal, N. & Lu, C. Identification of microRNAs and transcript targets in Camelina sativa by deep sequencing and computational methods. PLoS One 10(3), e0121542. https://doi.org/10.1371/journal.pone.0121542 (2015).
**ong, L. & Zhu, J.-K. Salt tolerance. Arabidopsis Book Am. Soc. Plant Biol. 1, 0048. https://doi.org/10.1199/tab.0048 (2002).
Machado, R. M. A. & Serralheiro, R. P. Soil salinity: Effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 3(30), 1–13 (2017).
Munns, R. & Gilliham, M. Salinity tolerance of crops—what is the cost?. New Phytol. 208, 668–673. https://doi.org/10.1111/nph.13519 (2015).
Sairam, R. K. & Tyagi, A. Physiology and molecular biology of salinity stress tolerance in plants. Curr. Sci. 86, 407–421 (2004).
Stearns, J. C., Woody, O. Z., McConkey, B. J. & Glick, B. R. Effects of bacterial ACC deaminase on Brassica napus gene expression. Mol. Plant Microbe Interact. 25(5), 668–676. https://doi.org/10.1094/MPMI-08-110213 (2012).
Heydarian, Z. et al. Changes in gene expression in Camelina sativa roots and vegetative tissues in response to salinity stress. Sci. Rep. 8, 9804. https://doi.org/10.1038/s41598-018-28204-4 (2018).
Ali, S., Charles, T. C. & Glick, B. R. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 80, 160–167 (2014).
Glick, B. R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. https://doi.org/10.1016/j.micres.2013.09.009 (2014).
Glick, B. R. Beneficial Plant–Bacterial Interactions 1–248 (Springer, Heidelberg, 2015).
Heydarian, Z. et al. Inoculation of soil with plant growth promoting bacteria producing 1-aminocyclopropane-1-carboxylate deaminase or expression of the corresponding acdS gene in transgenic plants increases salinity tolerance in Camelina sativa. Front. Microbiol. 7, 1966. https://doi.org/10.3389/fmicb.2016.01966 (2016).
Sergeeva, E., Shah, S. & Glick, B. R. Growth of transgenic canola (Brassica napus cv. Westar) expressing a bacterial1-aminocyclopropane-1-carboxylate (ACC) deaminase gene on high concentrations of salt. World J. Microbiol. Biotech. 22, 277–282 (2006).
Heydarian, Z., Gruber, M., Glick, B. R. & Hegedus, D. D. Gene expression patterns in roots of Camelina sativa with enhanced salinity tolerance arising from inoculation of soil with plant growth promoting bacteria producing 1-aminocyclopropane-1-carboxylate deaminase or expression the corresponding acdS gene. Front. Microbiol. 9, 1297. https://doi.org/10.3389/fmicb.2018.01297 (2018).
Rashid, S., Charles, T. C. & Glick, B. R. Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. J. 61, 217–224 (2012).
Duan, J., Jiang, W., Cheng, Z., Heikkila, J. J. & Glick, B. R. The complete genome sequence of the plant growth-promoting bacterium Pseudomonas putida UW4. PLoS One 8, e58640 (2013).
Ali, S., Charles, T. C. & Glick, B. R. Delay of flower senescence by bacterial endophytes expressing 1-aminocyclopropane-1-carboxylate deaminase. J. Appl. Microbiol. 113, 1139–1144 (2012).
Stringham, G. R. Genetics of four hypocotyl mutants in Brassica campestris L. J. Hered. 62(4), 248–250. https://doi.org/10.1093/oxfordjournals.jhered.a108161 (1971).
Wu, S., Feng, X. & Wittmeier, A. Microwave digestion of plant and grain reference materials in nitric acid or a mixture of nitric acid or a mixture of nitric acid and hydrogen peroxide for the determination of multi-elements by inductively coupled plasma mass spectrometry. J. Anal. Atmos. Spectrom. 12, 797–806. https://doi.org/10.1039/A607217H (1997).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29(1), 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Chen, D. et al. wheat aminocyclopropane-1-carboxylate oxidase gene, TaACO1, negatively regulates salinity stress in Arabidopsis thaliana. Plant Cell Rep. 33, 1815. https://doi.org/10.1007/s00299-014-1659-7 (2014).
Yaish, M. W. et al. A genome wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front. Plant Sci. 6, 946. https://doi.org/10.3389/fpls.2015.00946 (2015).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106. https://doi.org/10.1186/gb-2010-11-10-r106 (2010).
Klie, S. & Nikoloski, Z. The choice between mapman and gene ontology for automated gene function prediction in plant science. Front. Genet. 3, 115. https://doi.org/10.3389/fgene.2012.00115 (2012).
Feng, W., Lindner, H., Robbins, N. E. & Dinneny, J. R. Growing out of stress: The role of cell- and organ-scale growth control in plant water-stress response. Plant Cell. 28(8), 1769–1782. https://doi.org/10.1105/tpc.16.00182 (2016).
Lotkowska, M. E. et al. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 169(3), 1862–1880. https://doi.org/10.1104/pp.15.00605 (2015).
Morimoto, K. et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. 114, E8528–E8536. https://doi.org/10.1073/pnas.1704189114 (2017).
Stone, S. L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 5, 135. https://doi.org/10.3389/fpls.2014.00135 (2014).
Seo, D. et al. Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of ABA-mediated drought stress responses. Plant Phys. 160, 556–568. https://doi.org/10.1104/pp.112.202143 (2012).
Mertens, J., Aliyu, H. & Cowan, D. A. X. LEA proteins and the evolution of the WHy domain. Appl. Environ. Microbiol. 84(15), e00539-e618. https://doi.org/10.1128/AEM.00539-18 (2018).
Bao, Y. et al. Over expression of the NDR1/HIN1-Like gene NHL6 modifies seed germination in response to abscisic acid and abiotic stresses in Arabidopsis. PLoS One 11, e0148572. https://doi.org/10.1371/journal.pone.0148572 (2016).
Hanano, S. et al. A systematic survey in Arabidopsis thaliana of transcription factors that modulate circadian parameters. BMC Genom. 9, 182. https://doi.org/10.1186/1471-2164-9-182 (2008).
Gao, Q. M., Venugopal, S., Navarre, D. & Kachroo, A. Low 18:1-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476. https://doi.org/10.1104/pp.110.166876 (2010).
Phukan, U. J., Jeena, G. S. & Shukla, R. K. WRKY transcription factors: Molecular regulation and stress responses in plants. Fronti. Plant Sci. 7, 760. https://doi.org/10.3389/fpls.2016.00760 (2016).
Glick, B. R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica https://doi.org/10.6064/2012/963401 (2012).
Tsuchiya, T. et al. Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc Natl. Acad. Sci. 96(26), 15362–15367 (1999).
Zhu, X. G., Long, S. P. & Ort, D. R. X. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61, 235–261. https://doi.org/10.1146/annurev-arplant-042809-112206 (2010).
Kang, J. et al. Suppression of photosynthetic gene expression in roots is required for sustained root growth under phosphate deficiency. Plant Physiol. 165(3), 1156–1170. https://doi.org/10.1104/pp.114.238725 (2014).
Julkowska, M. M. & Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20(9), 586–594. https://doi.org/10.1016/j.tplants.2015.06.008 (2015).
Vaseva, I. I. et al. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. 115(17), E4130–E4139 (2018).
Achard, P. et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (2006).
Shen, X. et al. Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 86, 303–317. https://doi.org/10.1007/s11103-014-0230-9 (2014).
Lockhart, J. Salt of the Earth: Ethylene promotes salt tolerance by enhancing Na/K homeostasis. Plant Cell. 25(9), 3150. https://doi.org/10.1105/tpc.113.250911 (2013).
Tao, J. J. et al. The role of ethylene in plants under salinity stress. Front. Plant Sci. 6, 1059. https://doi.org/10.3389/fpls.2015.01059 (2015).
Cheng, Z., Woody, O. Z., McConkey, B. J. & Glick, B. R. Combined effects of the plant growth-promoting bacterium Pseudomonas putida UW4 and salinity stress on the Brassica napus proteome. Appl. Soil Ecol. 61, 255–263. https://doi.org/10.1016/j.apsoil.2011.10.006 (2012).
Zapata, P. J., Serrano, M., García-Legaz, M. F., Pretel, M. T. & Botella, M. A. Short term effect of salt shock on ethylene and polyamines depends on plant salt sensitivity. Front. Plant Sci. 8, 855. https://doi.org/10.3389/fpls.2017.00855 (2017).
Dong, H. et al. Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 62, 4875–4887. https://doi.org/10.1093/jxb/err143 (2011).
Dubois, M., den Broeck, L. V. & Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 4. https://doi.org/10.1016/j.tplants.2018.01.003 (2018).
Zhang, L. et al. An AP2 domain-containing gene ESE1 targeted by ethylene signaling component EIN3 is important for salt response in Arabidopsis thaliana. Plant Physiol. 157(2), 854–865. https://doi.org/10.1104/pp.111.179028 (2011).
Li, S., Fu, Q., Chen, L., Huang, W. & Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252 (2011).
Yang, Z., Tian, L., Latoszek-Green, M., Brown, D. & Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58(4), 585–596. https://doi.org/10.1007/s11103-005-7294-5 (2005).
Li, Z. G. et al. Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci. Rep. 24(5), 2477. https://doi.org/10.1038/srep12477 (2015).
Cohen, J. D. & Bandurski, R. S. Chemistry and physiology of the bound auxins. Annu. Rev. Plant Physiol. 33, 403–430 (1982).
Park, J. E. et al. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. Biol. Chem. 282, 10036–10046. https://doi.org/10.1074/jbc.M610524200 (2007).
LeClere, S., Tellez, R., Rampey, R. A., Matsuda, S. P. & Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277(23), 20446–20452 (2002).
Geng, Y. et al. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 25(6), 2132–2154. https://doi.org/10.1105/tpc.113.112896 (2013).
Gruner, K., Griebel, T., Návarová, H., Attaran, E. & Zeier, J. Reprogramming of plants during systemic acquired resistance. Front. Plant Sci. 4, 252. https://doi.org/10.3389/fpls.2013.00252 (2013).
Huot, B., Yao, J., Montgomery, B. L. & He, S. Y. Growth-defense trade offs in plants: A balancing act to optimize fitness. Mol. Plant. 7, 1267–1287. https://doi.org/10.1093/mp/ssu049 (2014).
Boestfleisch, C. & Papenbrock, J. Changes in secondary metabolites in the halophytic putative crop species Crithmum maritimum (L.), Triglochin maritima (L.) and Halimione portulacoides (L.) Aellen as reaction to mild salinity. PLoS One 12(4), 0176303. https://doi.org/10.1371/journal.pone.0176303 (2017).
Hegedus, D. D. et al. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397 (2003).
Podzimska-Sroka, D., O’Shea, C., Gregersen, P. L. & Skriver, K. NAC transcription factors in senescence: From molecular structure to function in crops. Plants 4, 412–448. https://doi.org/10.3390/plants4030412 (2015).
Kim, H. J. et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by Ethylene-Insensitive 2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036. https://doi.org/10.1093/jxb/eru112 (2014).
Msanne, J., Lin, J., Stone, J. M. & Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234(1), 97–107. https://doi.org/10.1007/s00425-011-1387-y (2011).
Li, S. et al. The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 35(8), 1729–1741. https://doi.org/10.1007/s00299-016-1991-1 (2016).
Peng, J. et al. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10(10), e1004664. https://doi.org/10.1371/journal.pgen.1004664 (2014).
Hickman, R. et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 75, 26–39. https://doi.org/10.1111/tpj.12194 (2013).
Fletcher, J. C. The Ultrapetala gene controls shoot and floral meristem size in Arabidopsis. Development 128, 1323–1333 (2001).
Fu, M., Kang, H. K., Son, S. H., Kim, S. K. & Nam, K. H. A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55(11), 1892–1904. https://doi.org/10.1093/pcp/pcu118 (2014).
Matías-Hernández, L., Aguilar-Jaramillo, A. E., Marin-Gonzalez, E., Suarez-Lopez, P. & Pelaz, S. RAV genes: Regulation of floral induction and beyond. Ann. Bot. 114, 1459–1470. https://doi.org/10.1093/aob/mcu069 (2014).
Guo, Y., Cai, Z. & Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549 (2004).
Miao, Y. & Zentgraf, U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. 19, 819–830 (2007).
Zhou, X., Jiang, Y. & Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells. 31, 303–313 (2011).
Besseau, S., Li, J. & Palva, E. T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63, 2667–2679 (2012).
Li, Z., Peng, J., Wen, X. & Guo, H. Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant. Biol. 54, 526–539 (2012).
Kodaira, K. S. et al. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 157, 742–756. https://doi.org/10.1104/pp.111.182683 (2011).
Acknowledgements
Z. Heydarian was a recipient of a Natural Sciences and Engineering Research Council Post-doctoral Fellowship from Agriculture and Agri-Food Canada.
Funding
This work was funded by a Grant from the Natural Resources Canada EcoEnergy Initiative.
Author information
Authors and Affiliations
Contributions
Z.H. conducted the research. Z.H. and C.C. conducted the informatics and statistical analysis. B.G. provided bacterial strains and the acdS gene. Z.H., M.G., B.G. and D.H. wrote the manuscript. M.G. and D.H. were the principal investigators and conceived of the research and obtained funding to support the project. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Heydarian, Z., Gruber, M., Coutu, C. et al. Gene expression patterns in shoots of Camelina sativa with enhanced salinity tolerance provided by plant growth promoting bacteria producing 1-aminocyclopropane-1-carboxylate deaminase or expression of the corresponding acdS gene. Sci Rep 11, 4260 (2021). https://doi.org/10.1038/s41598-021-83629-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-83629-8
- Springer Nature Limited
This article is cited by
-
Mitigating abiotic stress: microbiome engineering for improving agricultural production and environmental sustainability
Planta (2022)
-
A fruitful decade of bacterial ACC deaminase biotechnology: a pragmatic approach towards abiotic stress relief in plants
Theoretical and Experimental Plant Physiology (2022)