Abstract
The age-dependent decline in remyelination potential of the central nervous system during ageing is associated with a declined differentiation capacity of oligodendrocyte progenitor cells (OPCs). The molecular players that can enhance OPC differentiation or rejuvenate OPCs are unclear. Here we show that, in mouse OPCs, nuclear entry of SIRT2 is impaired and NAD+ levels are reduced during ageing. When we supplement β-nicotinamide mononucleotide (β-NMN), an NAD+ precursor, nuclear entry of SIRT2 in OPCs, OPC differentiation, and remyelination were rescued in aged animals. We show that the effects on myelination are mediated via the NAD+-SIRT2-H3K18Ac-ID4 axis, and SIRT2 is required for rejuvenating OPCs. Our results show that SIRT2 and NAD+ levels rescue the aged OPC differentiation potential to levels comparable to young age, providing potential targets to enhance remyelination during ageing.
Similar content being viewed by others
Introduction
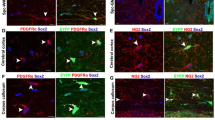
The increasingly ageing global population has ignited great interest in identifying rejuvenating molecules that may delay ageing and enhance the regeneration of the aged central nervous system (CNS). The ensheathment of axons by myelin gives structure to saltatory conduction1,2, and provides metabolic support to maintain both axonal functional integrity and long-term survival3,4. Therefore, proper myelination is not only a prerequisite for, but also a consequence of, normal CNS activity5,58. 70–100 normal myelinated (grade 0) axons were analysed per mouse. The myelin sheath was regarded as a torus, and the areas of the inner and outer circles of the myelin sheath were measured with a freehand tool on ImageJ by tracing the outer surface of each structure, then converted the areas into hypothetical radius (Supplementary Fig. 14). The G-Ratio was calculated as the ratio of the diameter of inner circle over that of the outer circle on the same myelin, which is inversely correlated with myelin thickness. The G-Ratio of unmyelinated and demyelinated axons is 1, which is the maximum value of the G-Ratio. The diameter of axons was calculated by area, which was also measured by tracking the outer surface of axons. Axon diameters were used in our scatter plot of axon diameter and G-Ratio, which represent the linear relationship between the G-Ratio and the axon diameter. The distance between DL was calculated by dividing the distance from the outermost DL to the innermost DL by the number of DL. In the mouse corpus callosum, the mean diameter of unmyelinated axons is more or less constant, with an overall mean diameter of 0.25 ± 0.01 μm59. Therefore, only axons with a diameter >0.25 μm were counted as unmyelinated axons. These axons are more likely to be axons without remyelination, rather than axons that were initially unmyelinated.
Evaluation of axonal pathology
“Defective axons” was evaluated by: shrunk axons, mitochondria accumulation or swelling, myelin sphere inclusion in axon or in the space between myelin and axon, and axon break. Only the axons with diameters >0.25 μm and were wrapped with myelin were analysed.
Immunopanning of OPCs
As previously described60, 50 μL BSL-1 (#L-1100, Vector Laboratories) was diluted in 22 mL DPBS, and four plates were coated with BSL-1 solution at RT for at least 4 h, then replaced by DPBS containing 0.2% BSA. Two 100 mm Petri dishes were incubated with 15 mL Tris buffer containing 72 µg goat anti-rat IgG (H + L) (#112-005-167, Jackson ImmunoResearch) at 4 °C overnight. Then, one of the secondary antibody-coated plates was washed three times with DPBS, and incubated with the primary antibody solution, which contained 10 μL rat anti-PDGFRα antibody (#558774, BD Biosciences-Pharmingen, 1:1000), 10 mL DPBS and 0.2% BSA, at RT for at least 2 h.
For tissue dissociation, 7-day-old WT and G3 Terc−/− mice brain tissues were dissected and incubated in the papain solution at 37 °C for 45–60 min. A trypsin inhibitor buffer was gently added to stop digestion. The dissociated cell solution was carefully layered over a 4 mL standard trypsin inhibitor buffer in a new 14 mL polystyrene tube. After being centrifuged at 200 g for 15 min, the cell pellet was resuspended with DPBS.
To deplete microglia, the cell suspension was incubated in a BSL-1 coated plate for 15 min at RT. Then, the supernatant was successively incubated in new BSL-1 coated plates for three times. The supernatant was transferred to a new tube, centrifuged at 200 g for 10 min and resuspended with a 6 mL panning buffer (DPBS, 0.02% BSA). To deplete the non-specific binding, cells were transferred to the secondary antibody-coated plate and incubated at RT for 8 min. Then the supernatant was transferred to the primary antibody-coated plate and incubated at RT for 15 min to collect OPCs. The floating cells were washed away with DPBS for 8–10 times. More washing should be performed if there is still significant number of floating cells. The remained cells were collected and stored at –80 °C until the next experiment.
Corpus callosum compound action potential (CAP) recording
As described previously, mice were first anesthetised with intraperitoneal injection of sodium pentobarbital (50 mg/kg) and decapitated. Then the brains were dissected out and transferred to an ice-cold slicing solution (2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 10 mM Dextrose, 213 mM Sucrose, 2 mM MgSO4, 2 mM CaCl2), which was bubbled with mixed gas (95% O2, 5% CO2). Coronal slices were cut (250 µm thick, four slices before and after bregma −1.0 mm) using a vibratome (Leica VT1200S). The slices were then transferred to an incubation chamber filled with artificial cerebrospinal fluid (ACSF) (126 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 25 mM Dextrose, 2 mM MgSO4, 2 mM CaCl2; 315–325 mOsm, pH = 7.2–7.3) and maintained at 34.5 °C for 45 min. After incubation, slices were kept in the same solution at RT and allowed to equilibrate at least 30 min prior to recording.
For electrophysiological recordings, Slices were transferred to a recording chamber perfused at 2 mL/min rate with aerated ACSF at 20.5–22.7 °C. Referring to a previous report40, a tungsten bipolar electrode was used for stimulation in the corpus callosum of one hemisphere and a glass electrode (impedance of 1–3 MΩ) filled with ACSF was placed in the contralateral hemisphere for recording. The stimulating pulses of 0.1 ms duration, 1 mA current and 10 s interval were applied via an isolator (ISO-Flex, A.M.P.I). Evoked CAPs were recorded by a MultiClamp 700B amplifier (Molecular Devices) and sampled by a Micro3 1401 (CED, Cambridge Electronic Design) at 25 kHz using Spike2 software (Bessel filter set to 10 kHz) for offline analysis. For each slice, we recorded two trials with the same stimulation site of two and different recording sites in the current clamp mode.
For data analysis and statistics, 100 repeated responses of each trial were averaged for waveform analysis. The time courses of conduction (T1 and T2) were calculated from the onset of stimulus artifact to negative peaks. Conduction velocity was estimated by the difference of two different distances between the stimulating and recording electrodes (δDistance) divided by the difference of corresponding peak latency (δT) in the same slice, that is, Fast velocity = δDistance/δT1; Slow velocity = δDistance/δT2. CAP amplitude was measured as the vertical distance from the local negative peak of two depolarising phases of the CAPs (Amp.1 and Amp.2) to a tangent joining preceding and following positivities. Due to the serious damage of myelin in some brain slices, the effective conduction was measured to reveal the percentage of slices which lack the fast component even with higher intensity of stimulus.
Targeted metabolic profiling
500 μL 80% Methanol (#67-56-1, Millipore) was added to OPCs (105 cells per sample) purified by immunopanning and vortexed fully to extract metabolites. 480 μL supernatant was collected after 15 min centrifugation (13000 g, 4 °C) and dried at SpeedVac Concentrator (ISS110-230, ThermoFisher) for 4 h. The dried metabolites were then reconstituted in 30 μL of 0.03% formic acid in analytical water. After being vortexed and centrifuged, the supernatant was transferred to an HPLC vial for metabolomics analysis. The targeted metabolite profiling was performed using an LC–MS/MS approach. Separation was achieved on an ACQUITY UPLC HSS T3 column (2.1 × 150 mm, 1.8 µm) using an Ultra High Performance Liquid Chromatograph (UHPLC) system (Nexera×2 LC-30A, Shimadzu) with the following gradient: 0–3 min, 1% mobile phase B; 3–15 min, 1–99% B; 15–17 min, 99% B; 17–17.1 min, 99–1% B; 17.1–20 min, 1% B. The mobile phases employed were 0.03% formic acid in water (A) and 0.03% formic acid in acetonitrile (B). The column was maintained at 35 °C and the samples were kept in the autosampler at 4 °C. The flow rate was 0.25 mL/min, and the injection volume was 20 μL. The mass spectrometer was a triple quadrupole mass spectrometer (QTRAP 6500 + , Sciex) with an electrospray ionisation (ESI) source. The sample analysis was performed in multiple reaction monitoring mode. A total of 256 metabolites were monitored with 160 ion transitions in positive mode and 96 ion transitions in negative mode. A chromatogram review and peak area integration were performed using the MultiQuant software v.3.0 (Sciex). The peak area for each detected metabolite was normalised against the total ion count of that sample, and thus is a fraction of the total detected metabolite content of that sample. Normalised peak areas were used as variables for multivariate and univariate statistical data analyses.
NAD+ quantification
NAD+ quantification was performed using NAD+/NADH Quantification Colorimetric Kit (#K337, BioVision) according to the instructions. Briefly, 20 mg brain tissues were washed with cold PBS and homogenised in 400 μL of NADH/NAD Extraction buffer in a micro-centrifuge tube, then centrifuged at 18000 g for 5 min and the supernatant transferred into a new tube and diluted 70 times with NADH/NAD Extraction buffer. For total NAD detection, 50 μL of diluted samples were transferred into a labeled 96-well plate. For NADH detection, the samples were incubated in a 60 °C water bath for 30 min to decompose the NAD+. For a standard curve preparation, 10 μL of 1 nmol/ul NADH standard was diluted with 990 μL NADH/NAD Extraction buffer to generate 10 pmol/μL standard NADH. Then 0, 2, 4, 6, 8, 10 μL of NADH standard were added into 96-well plate to generate a 0, 20, 40, 60, 80, 100 pmol/well standard. 100 μL of reaction mix (98 μL of NAD Cycling Buffer and 2 μL of NAD Cycling Enzyme Mix) was added into each well and incubated at RT for 5 min. Then 10 μL of NADH developer was added into each well and incubated at RT for 1–4 hours. The reactions were stopped by adding 10 μL of stop solution and the plate was read at OD 450 nm. The content of NAD+ was calculated by subtracting the content of NADH from total NAD.
Western blot analysis
For tissue lysis, the tissues had been ground in liquid nitrogen and lysed in lysis buffer [50 mM Tris-HCl-pH 7.5, 5 mM EDTA, 1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 3.69% CHAPS, 1% Triton X-100, 50 mM NaCl, 1 mM DTT (#3483-12-3, Biosharp), protease inhibitor (#18518900, Roche), phosphatase inhibitor A (#B15001-A, Biotool) and B (#B15001-B, Biotool), MG132] for 30 min on ice. Then, the mixture was centrifuged at 1000 g for 5 min at 4 °C and the supernatant was stored at –80 °C. For cell lysis, the cells were collected by centrifuged at 300 g for 5 min and lysed in lysis buffer at 4 °C for 30 min. For cellular fractionation, the cytoplasm and the nucleus of OLN93 cells were separated by using the “PARISTM kit” (ThermoFisher, #AM1921). Protein quantification was implemented using the BCA protein assay kit measured with a microplate reader. Equal amounts of protein (20–30 μg) were loaded mixed with 5X loading buffer (#FD006, Fdbio science) after boiling to 95 °C for 5 min. Proteins were loaded onto a 12% Blot Bis-Tris plus SDS gels (#FD346, Fdbio science) and run at 200 V in an SDS running buffer for 60 min for separation. Proteins were then transferred onto 0.45 μm PVDF membrane (#ISEQ00010, Millipore) in Blot transfer buffer at 300 mA for 75 min on ice. Blots were blocked at RT for 2 h in 5% skim milk blocking buffer in TBS-Tween (TBST). Primary antibodies were incubated overnight at 4 °C on a rocker in 0.1% TBST. Primary antibodies used in this study include: rabbit anti-SIRT2 (#S8447, Sigma, 1:1000), mouse anti-β-actin (#A5441, Sigma, 1:1000), mouse anti-Tubulin (#M1305-2, HUABIO, 1:5000), mouse anti-Ac-TubulinLys40 (#T7451, Sigma, 1:1000), rabbit anti-H3 (#ab1791, Abcam,1:10000), rabbit anti-H3K18Ac (#39694, Active motif, 1:1000), rabbit anti-H3K56Ac (#39282, Active motif, 1:1000), rat anti-MBP (#MCA409S, Bio-Rad Laboratories, 1:500), rabbit anti-ID4 (#LS-C384049, LSbio, 1:1000). Blots were washed with TBST (10 min, three times) and incubated at RT for 2 h with HRP secondary antibodies in TBST. Secondary antibodies used in this study include: Peroxidase affinipure donkey anti-rabbit IgG (H + L) (#711-035-152, Jackson ImmunoResearch, 1:10000), Peroxidase affinipure donkey anti-mouse IgG (H + L) (#715-035-151, Jackson ImmunoResearch, 1:10000). Blots were then washed with TBST (10 min, three times). HRP blots were developed with PierceTM ECL Western Blotting Substrate (#32106, Thermo Fisher Scientific) for 2 min and imaged with X-Ray films or ChemiDoc Touch Imaging Syetem (Bio-Rad). For the antibodies incubated in the same blots, after imaging, the blots were stripped with strip** buffer (25 mM Glycine and 1% SDS in ddH2O, pH 2.0) for 20 min at RT to remove antibodies and washed in TBST for 10 min three times. The blots were blocked at RT for 2 h in 5% skim milk blocking buffer in TBST and then incubated with another primary antibody. For protein quantification, densitometry was performed with ImageJ and normalised to β-actin.
Relative telomere length detection
Genomic DNA was extracted from 100 μL of venous blood which was drawn from the tail vein of mice using the TIANamp Genomic DNA Kit (#DP304, TIANGEN). Then DNA, with either telomere primers or 36B4 gene primers, and ChamQ Universal SYBR qPCR Master Mix (#Q711, Vazyme) were mixed as instructed by the manufacturer. Fold changes in gene expression were calculated using the delta Ct method in Microsoft Excel. Primer sequences: Tel-F, 5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′, Tel-R, 5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′, 36B4-F, 5′-CAGCAAGTGGGAAGGTGTAATCC-3′, 36B4-R: 5′-CCCATTCTATC ATCAACGGGTACAA-3′.
RNA isolation and qRT-PCR
RNA was isolated using the FastPure Cell/Tissue Total RNA Isolation Kit (#RC101, Vazyme). All RNA samples were stored at –80 °C until the next experiment. cDNA was generated using the HiScript III 1st Strand cDNA Synthesis Kit (#R312, Vazyme). For qRT-PCR, cDNA, primers and ChamQ Universal SYBR qPCR Master Mix (#Q711, Vazyme) were mixed as instructed by the manufacturer. Data were exported from the CFX Manager 3.0 and fold changes in gene expression were calculated using the delta-delta Ct method in Microsoft Excel. Primer sequences are listed in Supplementary Data 4.
Protein mass spectrometry
OPCs were lysed in RIPA (#P0013B, Beyotime) for 30 min on ice, then centrifuged at 1000 g for 5 min at 4 °C, and the supernatant was stored at –80 °C. The protein was quantified using a BCA protein assay kit (#P0011, Beyotime) measured with microplate reader (SynergyMx M5, Molecular Devices).
For protein precipitation and digestion, proteins were precipitated with acetone and the protein pellet was dried by using a Speedvac for 1−2 min. The pellet was subsequently dissolved in 8 M urea, 100 mM Tris-HCl, pH 8.5. TCEP (5 mM, Thermo Scientific) and iodoacetamide (10 mM, Sigma) for reduction and alkylation were added to the solution and incubated at RT for 30 min, respectively. The protein mixture was diluted four times and digested overnight with Trypsin at 1:50 (w/w) (Promega). The tryptic-digested peptide solution was desalted by using a MonoSpinTM C18 column (GL Science, Tokyo, Japan) and dried with a SpeedVac.
For LC/ tandem MS (MS/MS) analysis of peptide, the peptide mixture was injected twice as technical repeats and analysed by a home-made 35 cm-long pulled-tip analytical column (75 μm ID packed with ReproSil-Pur C18-AQ 1.9 μm resin, Dr. Maisch GmbH). The column was placed in-line with an Easy-nLC 1200 nano HPLC (Thermo Scientific, San Jose, CA) for mass spectrometry analysis. The analytical column temperature was set at 55 °C during the experiments. The mobile phase and elution gradient used for peptide separation were as follows: 0.1% formic acid in water as buffer A and 0.1% formic acid in 80% acetonitrile as buffer B, 0–1 min, 3–8% B; 1–191 min, 8–25% B; 191–219 min, 25–50% B, 219–220 min, 50–100% B, 220–240 min, 100% B. The flow rate was set as 300 nL/min.
For Mass Spectrometry, data-dependent tandem mass spectrometry (MS/MS) analysis was performed with a Q Exactive Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA). Peptides eluted from the LC column were directly electrosprayed into the mass spectrometer with the application of a distal 2 kV spray voltage. A cycle of one full-scan MS spectrum (m/z 300–1800) was acquired followed by top 20 MS/MS events, sequentially generated on the first to the twentieth most intense ions selected from the full MS spectrum at a 28% normalised collision energy. The full-scan resolution was set to 70,000 with automated gain control (AGC) target of 3e6. The MS/MS scan resolution was set to 17,500 with an isolation window of 1.8 m/z and AGC target of 1e5. The number of microscans was one for both MS and MS/MS scans and the time of maximum ion injection was 50 and 100 ms, respectively. The dynamic exclusion settings used were as follows: charge exclusion, 1 and >8; exclude isotopes, on; and exclusion duration, 30 s. MS scan functions and LC solvent gradients were controlled by the Xcalibur data system (Thermo Scientific).
For data analysis, the acquired MS/MS data were analysed against the UniProt Knowledgebase (https://www.uniprot.org, Swiss-Prot Rattus norvegicus database released on Apr. 1, 2019) using PEAKS Studio 8.5 (Bioinformatics Solutions, Waterloo, Ontario, Canada). The database search parameters were set as the followings: MS and MS/MS tolerance of 20 ppm and 0.1 Da, respectively, FDR was set as 1% and the protein identification threshold was set as (−10 logP) ≧ 20. A protein was identified when at least one unique peptide was matched. Protein quantification was based on label-free quantitative analysis.
Lentiviral constructs and production
We initially developed the lentiviral transfer plasmid. For SIRT2 over expression, we inserted the sirt2 gene into the pCDH-CMV-EF1α-flag-P2A-copGFP plasmid. For SIRT2 knockdown, we designed shRNA sequences using the BLOCK-iTTM RNAi Designer (Thermo Fisher) and inserted the sequences into the pGreenPuro plasmid (#SI505A-1, System Biosciences). Then the lentivirus producer cell line HEK239T was transiently co-transfected with transfer (pCDH-CMV-EF1α-SIRT2-flag-P2A-copGFP or pGreenPuro-shSIRT2 plasmid), envelope (pMD2.G, #12259, Addgene) and packaging (psPAX2, #12260, Addgene) plasmids to generate lentiviral particles. HEK293T cells were transfected using Polyethylenimine (PEI, #23966, Polysciences Inc., 1 mg/mL). After 8 h of transfection, the medium was changed to a fresh medium, and the lentiviral was collected after 48 h of transfection by centrifuged at 800 g at 4 °C for 10 min to remove cell debris. The SIRT2 shRNA sequences are: shRNA-F, 5′-gatccGGATGAAAGAGAAGATCTTCTttcaagagaAGAAGATCTTCTCTTTCATCCTTTTTg-3′, shRNA-R, 5′-aattcAAAAAGGATGAAAGAGAAGATCTTCTtctcttgaaAGAAGATCTTCTCTTTCATCCg-3′.
Construction of stable cell lines
5 × 105 OLN93 cells per well were seeded into a six-well plate and the culture medium was replaced with the collected lentiviral supernatant after 12 h. Fluorescence was observed after 48 h of lentivirus infection. The cells were gradually diluted and seeded into a 96-well plate and fluorescent cell clones were selected for passage after 2 weeks of culture.
ChIP-qPCR
For the cross-linking of protein and DNA, 270 μL of 37% formaldehyde was added into a 10 cm dish when the cell confluency reaches 90%, and the dish was shaken slowly for 10 min at RT. Then 500 μL of 2.5 M glycine was added into the medium and the dish was shaken for 5 min to quench cross-linking. The cells were washed three times with cold PBS and digested in 0.25% trypsin for 3 min, then collected by centrifugation (800 g for 5 min at 4 °C) and washed twice with cold PBS. Cells were lysed in 500 μL ChIP-lysis buffer (50 mM Tris-HCl-pH 8.0, 0.5% SDS, 5 mM EDTA, protease inhibitor) for 5 min at 4 °C and centrifugated briefly. The samples were sonicated to shear the chromatin into about 300 bp fragments (typically, 0.5 s pulses followed by 0.5 s rest periods for 15 min at output 10). Then 5–10 μg of antibody or IgG was added into samples and incubated overnight at 4 °C. Antibodies used in this study include: Rabbit anti-SIRT2 (#S8447, Sigma, 1:100), mouse anti-flag (#M185-3L, MBL, 1:100), rabbit anti-H3K18Ac (#39694, Active motif), normal mouse IgG (#A7028, Beyotime), and normal rabbit IgG (#A7016, Beyotime). A 1% volume of the sample was pipetted out as input and the rest sample was centrifugated and the supernatant was transferred to a fresh micro-centrifuge tube. 50 μL Protein A/G PLUS-Agarose (#sc-2003, SANTA CRUZ) was blocked in the block buffer (10 mM Tris-HCl-pH 8.0, 1 mM EDTA-pH 8.0, 1% BSA) twice for 5 min and centrifugated for 2 min (800 g, 4 °C). The samples were incubated with agarose for 2 h at 4 °C and centrifugated for 2 min (800 g, 4 °C) and then the samples were washed with wash buffer I (20 mM Tris-HCl-pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% TritonX-100, 0.1% SDS), wash buffer II (20 mM Tris-HCl-pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% TritonX-100, 0.1% SDS), wash buffer III (10 mM Tris-HCl-pH 8.0, 0.25 M LiCl, 1 mM EDTA, 1% NP-40), TE buffer (10 mM Tris-HCl-pH 8.0, 1 mM EDTA-pH 8.0), TE buffer for 10 min at 4 °C followed by 2 min of centrifugation (800 g, 4 °C), respectively. The agarose was resuspended with 100 μL elution buffer (0.1 M NaHCO3, 1% SDS, 1 mg/mL protease K) twice by using a rotary shaker for 10 min at RT and the supernatant was collected after centrifugation (800 g, 2 min, RT). The sample tubes were placed in a 65 °C water bath overnight to release cross-linking. The DNA was purified from samples using a Universal DNA Purification Kit (#DP214, TIANGEN) as instructed by the manufacturer.
For qPCR, DNA, primers and ChamQ Universal SYBR qPCR Master Mix (#Q711, Vazyme) were mixed as instructed by the manufacturer. Fold changes were calculated using the Delta-Delta Ct method in Microsoft Excel. The primer sequences are as follows: ChIP-ID4-F, 5′-TGGCACTGTCCTCCTGATTG-3′, ChIP-ID4-R, 5′-CCCTCAAAGTAACGACTTCCAA-3′.
Statistics and reproducibility
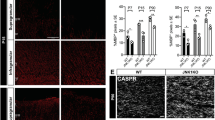
All data analyses were performed using GraphPad Prism 8.0 or R. The data are shown as mean ± SEM. All the images, immunoblots and statistical graphs are representatives of at least three experiments, unless otherwise stated. The “n” numbers for each experiment are specified in the figure legends. For in vivo experiments, each data point represents an individual animal. For in vitro experiments, each data point represents a biological replicate. For the comparison between two groups, statistical significance was determined using the unpaired two-tailed Student’s t test. For multiple comparisons, a one-way ANOVA test, followed by Tukey’s post hoc test, or two-way ANOVA, followed by Sidak’s multiple comparisons, was performed. Differences were considered statistically significant if p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***). The heatmap was generated with the ggplot2 library.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data was analysed against the UniProt Knowledgebase (https://www.uniprot.org, Swiss-Prot Rattus norvegicus database released on Apr. 1 st 2019). The mass spectrometry proteomics raw data generated during this study is available at the ProteomeXchange Consortium via the iProX partner repository, with the dataset identifier PXD022046 and IPX0002529000. All other relevant data supporting the key findings of this study is available within the article and its Supplementary Information files. The source data generated in this study is provided in the Source Data files. Source data are provided with this paper.
References
Tasaki, I. The electro-saltatory transmission of the nerve impulse and the effect of narcosis upon the nerve fiber. Am. J. Physiol. 127, 211–227 (1939).
Huxley, A. F. & Stampfli, R. Evidence for saltatory conduction in peripheral myelinated nerve fibres. J. Physiol. 108, 315–339 (1949).
Brown, A. M., Evans, R. D., Black, J. & Ransom, B. R. Schwann cell glycogen selectively supports myelinated axon function. Ann. Neurol. 72, 406–418 (2012).
Funfschilling, U. et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521 (2012).
McKenzie, I. A. et al. Motor skill learning requires active central myelination. Science 346, 318–322 (2014).
**ao, L. et al. Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nat. Neurosci. 19, 1210–1217 (2016).
Meier-Ruge, W., Ulrich, J., Bruhlmann, M. & Meier, E. Age-related white matter atrophy in the human brain. Ann. N. Y Acad. Sci. 673, 260–269 (1992).
Sturrock, R. R. Changes in neurologia and myelination in the white matter of aging mice. J. Gerontol. 31, 513–522 (1976).
**e, F., Liang, P., Fu, H., Zhang, J. C. & Chen, J. Effects of normal aging on myelin sheath ultrastructures in the somatic sensorimotor system of rats. Mol. Med Rep. 10, 459–466 (2014).
Peters, A. The effects of normal aging on myelin and nerve fibers: a review. J. Neurocytol. 31, 581–593 (2002).
Chapman, T. W. & Hill, R. A. Myelin plasticity in adulthood and aging. Neurosci. Lett. 715, 134645 (2020).
Franklin, R. J. M. & Ffrench-Constant, C. Regenerating CNS myelin - from mechanisms to experimental medicines. Nat. Rev. Neurosci. 18, 753–769 (2017).
Kang, S. H. et al. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat. Neurosci. 16, 571–579 (2013).
Dean, D. C. 3rd et al. Association of Amyloid Pathology With Myelin Alteration in Preclinical Alzheimer Disease. JAMA Neurol. 74, 41–49 (2017).
Mei, F. et al. Accelerated remyelination during inflammatory demyelination prevents axonal loss and improves functional recovery. Elife 5, e18246 (2016).
Hill, R. A., Li, A. M. & Grutzendler, J. Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat. Neurosci. 21, 683–695 (2018).
Cantuti-Castelvetri, L. et al. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science 359, 684–688 (2018).
Neumann, B. et al. Metformin Restores CNS Remyelination Capacity by Rejuvenating Aged Stem Cells. Cell Stem Cell 25, 473–485 e478 (2019).
Rawji, K. S. et al. Niacin-mediated rejuvenation of macrophage/microglia enhances remyelination of the aging central nervous system. Acta Neuropathol. 139, 893–909 (2020).
Ruckh, J. M. et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell 10, 96–103 (2012).
Shen, S. et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat. Neurosci. 11, 1024–1034 (2008).
Thompson, A. J., Baranzini, S. E., Geurts, J., Hemmer, B. & Ciccarelli, O. Multiple sclerosis. Lancet 391, 1622–1636 (2018).
Neumann, B., Segel, M., Chalut, K. J. & Franklin, R. J. Remyelination and ageing: Reversing the ravages of time. Mult. Scler. 25, 1835–1841 (2019).
Zhao, J. W., Raha-Chowdhury, R., Fawcett, J. W. & Watts, C. Astrocytes and oligodendrocytes can be generated from NG2+ progenitors after acute brain injury: intracellular localization of oligodendrocyte transcription factor 2 is associated with their fate choice. Eur. J. Neurosci. 29, 1853–1869 (2009).
Ren, R., Ocampo, A., Liu, G. H. & Izpisua Belmonte, J. C. Regulation of Stem Cell Aging by Metabolism and Epigenetics. Cell Metab. 26, 460–474 (2017).
Wang, Y., Yang, J., Hong, T., Chen, X. & Cui, L. SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res Rev. 55, 100961 (2019).
Gomes, P., Fleming Outeiro, T. & Cavadas, C. Emerging Role of Sirtuin 2 in the Regulation of Mammalian Metabolism. Trends Pharm. Sci. 36, 756–768 (2015).
Foolad, F., Khodagholi, F. & Javan, M. Sirtuins in Multiple Sclerosis: the crossroad of neurodegeneration, autoimmunity and metabolism. Mult. Scler. Relat. Disord. 34, 47–58 (2019).
Li, W. et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J. Neurosci. 27, 2606–2616 (2007).
Werner, H. B. et al. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J. Neurosci. 27, 7717–7730 (2007).
Ji, S., Doucette, J. R. & Nazarali, A. J. Sirt2 is a novel in vivo downstream target of Nkx2.2 and enhances oligodendroglial cell differentiation. J. Mol. Cell Biol. 3, 351–359 (2011).
Beyer, B. A. et al. Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat. Chem. Biol. 14, 22–28 (2018).
Bercury, K. K. & Macklin, W. B. Dynamics and mechanisms of CNS myelination. Dev. Cell 32, 447–458 (2015).
Zhu, X. et al. Age-dependent fate and lineage restriction of single NG2 cells. Development 138, 745–753 (2011).
McLane, L. E. et al. Loss of Tuberous Sclerosis Complex1 in Adult Oligodendrocyte Progenitor Cells Enhances Axon Remyelination and Increases Myelin Thickness after a Focal Demyelination. J. Neurosci. 37, 7534–7546 (2017).
Kosaraju, J. et al. Metformin promotes CNS remyelination and improves social interaction following focal demyelination through CBP Ser436 phosphorylation. Exp. Neurol. 334, 113454 (2020).
Lasiene, J., Matsui, A., Sawa, Y., Wong, F. & Horner, P. J. Age-related myelin dynamics revealed by increased oligodendrogenesis and short internodes. Aging Cell 8, 201–213 (2009).
Martinez, P. & Blasco, M. A. Telomere-driven diseases and telomere-targeting therapies. J. Cell Biol. 216, 875–887 (2017).
Tacutu, R. et al. Human Ageing Genomic Resources: new and updated databases. Nucleic Acids Res. 46, D1083–D1090 (2018).
Crawford, D. K., Mangiardi, M. & Tiwari-Woodruff, S. K. Assaying the functional effects of demyelination and remyelination: revisiting field potential recordings. J. Neurosci. Methods 182, 25–33 (2009).
Das, C., Lucia, M. S., Hansen, K. C. & Tyler, J. K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009).
Eskandarian, H. A. et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 341, 1238858 (2013).
Kondo, T. & Raff, M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J. 19, 1998–2007 (2000).
Marin-Husstege, M. et al. Multiple roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia 54, 285–296 (2006).
Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Plemel, J. R., Liu, W. Q. & Yong, V. W. Remyelination therapies: a new direction and challenge in multiple sclerosis. Nat. Rev. Drug Disco. 16, 617–634 (2017).
Lovato, L. et al. Transketolase and 2’,3’-cyclic-nucleotide 3’-phosphodiesterase type I isoforms are specifically recognized by IgG autoantibodies in multiple sclerosis patients. Mol. Cell Proteom. 7, 2337–2349 (2008).
Jastorff, A. M. et al. Regulation of proteins mediating neurodegeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. Proteom. Clin. Appl. 3, 1273–1287 (2009).
Lautrup, S., Sinclair, D. A., Mattson, M. P. & Fang, E. F. NAD(+) in Brain Aging and Neurodegenerative Disorders. Cell Metab. 30, 630–655 (2019).
Covarrubias, A. J., Perrone, R., Grozio, A. & Verdin, E. NAD(+) metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 22, 119–141 (2021).
Irie, J. et al. Effect of oral administration of nicotinamide mononucleotide on clinical parameters and nicotinamide metabolite levels in healthy Japanese men. Endocr. J. 67, 153–160 (2020).
Gokhan, S. et al. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J. Neurosci. 25, 8311–8321 (2005).
Wang, J. et al. Treatment with NAD(+) inhibited experimental autoimmune encephalomyelitis by activating AMPK/SIRT1 signaling pathway and modulating Th1/Th17 immune responses in mice. Int. Immunopharmacol. 39, 287–294 (2016).
Tullius, S. G. et al. NAD+ protects against EAE by regulating CD4+ T-cell differentiation. Nat. Commun. 5, 5101 (2014).
Diao, D. et al. Telomeric epigenetic response mediated by Gadd45a regulates stem cell aging and lifespan. EMBO Rep. 19, e45494 (2018).
Tang, X. et al. SIRT2 Acts as a Cardioprotective Deacetylase in Pathological Cardiac Hypertrophy. Circulation 136, 2051–2067 (2017).
Goebbels, S. et al. Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrap** and myelination. J. Neurosci. 30, 8953–8964 (2010).
Sturrock, R. R. Myelination of the mouse corpus callosum. Neuropathol. Appl. Neurobiol. 6, 415–420 (1980).
Emery, B. & Dugas, J. C. Purification of oligodendrocyte lineage cells from mouse cortices by immunopanning. Cold Spring Harb. Protoc. 2013, 854–868 (2013).
Acknowledgements
We thank the Center of Cryo-Electron Microscopy, Zhejiang University (ZJU), and the Core Facilities of ZJU School of Medicine. Particular thanks to Drs. ** Hu
Contributions
J.-W.Z. and Z.J. conceived the project; J.-W.Z., Z.J., X.-R.M., and X.Z. designed the experiment; X.-R.M., X.Z., Y.X., H.-M.G., S.-S.Z., Liang Li, C.Y., W.J., K.Y., Y.Y., and C.P. performed most of the experiments and acquired data; X.-R.M., Y.X., H.-M.G., S.-S.Z., Liang Li, W.J., C.L., K.Y., Z.H., Z.M., and Y.Y. analysed and interpreted the data; F.W., Z.-J.D., D.-X.W., Y.W., Y.Z. and L.G. assisted with the experiments and helped to analyse the data. J.-W.Z. and X.-R.M. wrote the paper; Y.S., H.-Z.C., X.Z. and Li Li edited the paper; revised the paper. J.-W.Z. supervised all phases of the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Danica Chen and the other anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, XR., Zhu, X., **ao, Y. et al. Restoring nuclear entry of Sirtuin 2 in oligodendrocyte progenitor cells promotes remyelination during ageing. Nat Commun 13, 1225 (2022). https://doi.org/10.1038/s41467-022-28844-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-28844-1
- Springer Nature Limited
This article is cited by
-
Sirtuins in kidney diseases: potential mechanism and therapeutic targets
Cell Communication and Signaling (2024)
-
Focusing on mitochondria in the brain: from biology to therapeutics
Translational Neurodegeneration (2024)
-
Repression of developmental transcription factor networks triggers aging-associated gene expression in human glial progenitor cells
Nature Communications (2024)
-
Targeting NAD Metabolism for the Therapy of Age-Related Neurodegenerative Diseases
Neuroscience Bulletin (2024)
-
GDF11 slows excitatory neuronal senescence and brain ageing by repressing p21
Nature Communications (2023)