Abstract
Triple-negative breast cancer (TNBC) is the most aggressive subtype of mammary carcinoma. Here, we describe a case of an 81-year-old female diagnosed with ductal triple negative breast cancer with a germline pathogenic variant in BReast CAncer gene2 (BRCA2). Genetic testing also revealed the presence of four somatic mutations in the ephrin type-A receptor 3 (EphA3), TP53, BRCA1-associated protein (BAP1), and MYB genes. The BRCA2, TP53, and BAP1 gene mutations are highly predictive of a defective homologous recombination repair system and subsequent chromosomal instability in this patient. Coherently, the patient displayed a strong homologous recombination deficiency signature and high tumor mutational burden status, which are generally associated with increased probability of immune neoantigens formation and presentation, and with tumor immunogenicity. Analysis of immune checkpoint revealed high expression of programmed cell death ligand 1 (PD-L1), programmed cell death ligand 2 (PD-L2), programmed death 1 (PD1), and cytotoxic T-lymphocyte-associated protein 4 (CTLA 4), suggesting that the patient might likely benefit from immunotherapies. Altogether, these findings support an unveiled link between BRCA2 inactivation, HR deficiency and increased expression of immune checkpoints in TNBC. This clinical case highlights the importance of screening TNBC patients for genetic mutations and TMB biomarkers in order to predict the potential efficacy of immunotherapy.
Similar content being viewed by others
Introduction
Breast cancer is one of the most common malignancies in women worldwide [1]. It displays complex diversity in both molecular alterations, clinical manifestations, and pathological characteristics [2,3,4,5]. Estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (ERBB2/HER2) are considered as the molecular markers for diagnostic classification of breast cancer subtypes [6, 7]. Breast cancer with the genetic signature of ER-negative, PR-negative, and ERBB2/HER2-negative has been classified as triple-negative breast cancer (TNBC) [8], which represents the most aggressive clinical subtype with a poor prognosis. Breast cancer, and in particular TNBC, presents significant genomic defects [9,10,11,12] involving protein degradation [13,14,15], mutations or deregulation of the p53 family members of tumor suppressors [16,17,18,19,20] as well as of other transcription factors [21,22,23]. In addition, also defects in metabolism or in hypoxia response [24,25,52,53,54,55]. As shown in Fig. 1G, the patient indeed showed absence of ER1, ER2, PR and HER2 mRNAs as compared to the clinical cohort (580 breast cancer patients). Further characterization of the tumor revealed a basal subtype of TNBC characterized by low to absent luminal differentiation marker expression, and high expression of epithelial-to-mesenchymal transition (EMT) and cancer stem cell-like markers (e.g., low claudin). The patient underwent a comprehensive genomic profiling that indicated the presence of four concurrent heterozygous somatic mutations in the ephrin type-A receptor 3 (EphA3), TP53, BRCA1-associated protein (BAP1) and MYB genes (Table 2).
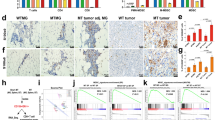
A Hematoxylin and eosin staining shows a G3 infiltrating ductal carcinoma with inflammatory infiltrates. Scale bar represents 50 µm. B High magnification of panel A highlights the presence of peritumoral inflammatory cells (asterisk). Scale bar represents 50 µm. C Ki67 expression in more than 70% of breast cancer cells. Scale bar represents 50 µm. D c-Erb-B2 staining revealed score 0. Scale bar represents 20 µm. E PDL-1 (SP142) immunostaining shows positivity in more than 1% of tumor-associated lymphocytes (asterisk). Scale bar represents 100 µm. F High magnification of panel (E). scale bar represents 20 µm. G Expression mRNA levels (TPM) of estrogen receptor 1 (ESR1), progesterone receptor (PR1), ERBB2 receptor tyrosine kinase 2 (HER2) and proliferation marker KI-67 (MKI67) for the patient (red triangle) and the clinical cohort (blue boxplot).
The His214 frameshift mutation of TP53 (Table 2) lies within its DNA-binding domain (DBD), suggesting that it may impair its ability to contact DNA (Fig. 2A). This mutation has not been described previously in cancer patients. Interestingly, Yaupt and co-authors reported a truncated protein (p53d1214), composed by the first 214 amino-terminal residues of p53 that lacks the transactivation function, even though retains the ability to induce apoptosis [41, 56]. The relevance of this mutation in vivo and its biological significance warrants further studies.
A Schematic structure of the p53 protein and lollipop plot showing the incidence of mutations in the TP53 gene in METABRIC cohort. Patient’s mutation is indicated. B Schematic structural features of the BAP1 protein and lollipop plot showing the incidence of BAP1 gene mutations in METABRIC cohort. Patient’s mutation is indicated by arrow. C Schematic structural features of the BRCA2 protein and lollipop plot showing incidence of mutations in the BRCA2 gene in METABRIC cohort. Patient’s mutation is indicated by arrow. A–C Data were obtained from cBioPortal. D Mutational contribution of HR-related signatures. E The patient has a higher TMB as compared to the cohort median (~80% percentile). F MSI score (MSI High: score > 0.901). The patient is observed as having MSI Low status (score = 0.09). G Chromosomal instability: CNH is higher in the patient compared to the median disease cohort, whereas numerical and structural CIN values are not much higher than median. The patient (red triangle) is compared to the clinical cohort (blue boxplot).
We also identified a somatic mutation in the BAP1 gene encoding a ubiquitin carboxy-terminal hydrolase that regulates several important cellular responses including HR DNA repair and cell growth [57, 58]. Germline inactivation of BAP1 confers an increased risk for develo** cutaneous and uveal melanoma, mesothelioma, renal cell carcinoma, and breast cancer, albeit to a lesser extent [59]. Mutations of BAP1 have been included in the HR-deficiency-associated pathways in breast cancer, particularly in the TNBC subtype, characterized by a relative high mutation frequency [85], implying that the patient presented in this study would be probably sensitive to this treatment.
Methods
Collection of samples
Tumor tissues were globally collected using a standardized protocol, minimizing the ischemia time until freezing in liquid nitrogen. To ensure the quality of the samples, all tissues were Hematoxilin and Eosin stained and subjected to a pathological QC. Samples need to be invasive, have a tumor content of ≥30% and Necrosis ≤30%. Normal tissues were processed in parallel and need to be free of tumor and representative regarding the tumor tissue to be included.
Approximately 10 mg tissue were taken for nucleic acid extraction and protein lysate preparation each. To account for tumor heterogeneity, pathological QCs were performed on two sections, before and after taking the analysis material. The tissues stay frozen during the entire process.
Immunohistochemical analysis
Approximately 1 × 1 × 0.5 cm of tissue was formalin-fixed and paraffin-embedded (FFPE). Serial sections were used to evaluate prognostic and predictive biomarkers including ER, PR, Ki67, and HER2 through immunohistochemistry. Briefly, sections were stained using the automated Leica Bond IHC platform (Leica Biosystems, Deer Park, IL). After antigen retrieval, 4-μm thick sections were incubated with the following primary monoclonal antibodies: mouse monoclonal anti-Ki67 (clone MM1; Leica Biosystems), mouse monoclonal anti-HER2 (clone CB11, Leica Biosystems) and rabbit monoclonal anti-PDL1 (clone sp142; Ventana Roche, USA). Reactions were revealed using BOND-PRIME Polymer DAB Detection System (Leica Biosystems, Deer Park, IL). Immunohistochemistry was evaluated by two blind pathologists.
Nucleic acid extraction and quality assessment
Frozen tissue slices were mixed with beta-mercaptoethanol containing sample buffer and homogenized using the BeadBug system. DNA and RNA were extracted in parallel from the same sample using the Qiagen AllPrep Universal Kit according to the manufacturer’s instructions.
DNA and RNA concentration were quantified using Qubit fluorometer with the Qubit dsDNA BR assay or Qubit RNA BR assay respectively.
DNA and RNA quality were assessed using the Agilent Tapestation with the Agilent Genomic DNA kit or Agilent High-Sensitivity RNA ScreenTape kit respectively. RNAs need to have a RIN ≥ 4 or a DV200 ≥ 60 to be selected for library preparation.
Library preparation and NGS sequencing
Libraries for whole genome sequencing (WGS) were prepared using the PCR-free KAPA Hyper Prep Kit (Roche). For whole transcriptome sequencing, RNA samples were depleted of the ribosomal RNA using the Ribo Zero Kit (Illumina) and library preparation was performed using the TruSeq Stranded Total RNA Kit (Qiagen). For small RNA sequencing the QIAseq miRNA Kit (Qiagen) was used All library preparation kits were used according to manufacturer’s instructions. Sequencing was performed on a NovaSeq6000 system (Illumina).
For WGS, average coverage for tumor samples was ≥60X and ≥30X for normal samples with a total genomic coverage of ≥95%.
Whole transcriptome sequencing datasets have ≥100 million total reads with <20% of ribosomal origin and ≥20 million reads map** to mRNAs according to Ensembl reference. Ribosomal depletion was performed to remove nuclear rRNA and mt-rRNA.
NGS data processing
NGS data was aligned against Grch38 genome assembly. Identification and annotation of short genomic variations in normal sample was done using Haplotype Caller (genome analysis toolkit; GATK) [86]. WGS somatic variation were called using a consensus of Mutect2(ref. 87), Strelka [88], Varscan [89] and Somatic Sniper [90]. Structural variations were called using R packages TitanCNA [91]and DellyCNV [92].
RNA-Seq differential expression was based on normalized readcount data (TPM: transcripts per million).
Mass spectrometry phospho-proteome profiling
For phospho-proteome profiling, 5–10 mg of fresh-frozen tissue was lysed in 2 mL Precellys® CK14 tubes containing 1.4 mm ceramic beads and using a lysis buffer containing PhosSTOP™ and bead shaking using a Precellys® Evolution Homogenizer equipped with a Cryolys® cooling module. After overnight digest samples were acidified and subjected to peptide desalting using Waters HLB Oasis 30 mg 96-well plates. 500 μg of peptide preparation was subjected to phospho-peptide enrichment using MagReSyn® Ti-IMAC magnetic beads (ReSyn Biosciences) as described in ref. 93 with modifications to enable processing using a KingFisher™ Flex robot equipped with a 96-magnetic pin head. Peptides were desalted using Waters μElution plates, dried down and resolubilized.
For DIA LC-MS/MS measurements, 5 μg of peptides per sample were injected to a reversed phase column (nanoEase M/Z Peptide CSH C18 Column, 1.7 μm, 300 μm X 150 mm) on a Waters ACQUITY UPLC M-Class LC connected to a Thermo Scientific™ Orbitrap Q Exactive™ HF-X mass spectrometer equipped with an EASYspray source. The nonlinear LC gradient was 1–60 % solvent B in 60 min at 50 °C and a flow rate of 5 μL/min. The DIA method consisting of one full range MS1 scan and 50 DIA segments was adapted from Bruderer et al. [94].
Tissue-specific spectral libraries were generated combining high-fractionated DDA and DIA measurements on a pool of tissue material and raw data processed using Biognosys’ software Spectronaut 13.
Bioinformatical analyses
Mutational signatures were calculated using the R package MutationalPatterns [95]. MSI classification was done using R package MSIseq [96]. PAM50 subty** as well as risk scores were investigated using R package genefu [97].
TMB was calculated as the number of non-synonymous mutations of protein-coding genes divided by exome size in Megabases.
References
Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radio. 2022;95:20211033.
Scimeca M, Urbano N, Bonfiglio R, Schillaci O, Bonanno E. Breast osteoblast-like cells: a new biomarker for the management of breast cancer. Br J Cancer. 2018;119:1129–32.
Scimeca M, Bonfiglio R, Menichini E, Albonici L, Urbano N, De Caro MT, et al. Microcalcifications drive breast cancer occurrence and development by macrophage-mediated epithelial to mesenchymal transition. Int J Mol Sci. 2019;20:5633.
Scimeca M, Trivigno D, Bonfiglio R, Ciuffa S, Urbano N, Schillaci O, et al. Breast cancer metastasis to bone: from epithelial to mesenchymal transition to breast osteoblast-like cells. Semin Cancer Biol. 2021;72:155–64.
Scimeca M, Giocondo R, Montanaro M, Granaglia A, Bonfiglio R, Tancredi V, et al. BMP-2 variants in breast epithelial to mesenchymal transition and microcalcifications origin. Cells. 2020;9:1381.
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013.
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95.
Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389:2430–42.
Marchetti P, Antonov A, Anemona L, Vangapandou C, Montanaro M, Botticelli A, et al. New immunological potential markers for triple negative breast cancer: IL18R1, CD53, TRIM, Jaw1, LTB, PTPRCAP. Discov Oncol. 2021;12:6.
Romera-Giner S, Andreu Martínez Z, García-García F, Hidalgo MR. Common pathways and functional profiles reveal underlying patterns in Breast, Kidney and Lung cancers. Biol Direct. 2021;16:9.
Amelio I, Bertolo R, Bove P, Candi E, Chiocchi M, Cipriani C, et al. Cancer predictive studies. Biol Direct. 2020;15:18.
Ganini C, Amelio I, Bertolo R, Bove P, Buonomo OC, Candi E, et al. Global map** of cancers: The Cancer Genome Atlas and beyond. Mol Oncol. 2021;15:2823–40.
Ren C, Han X, Lu C, Yang T, Qiao P, Sun Y, et al. Ubiquitination of NF-κB p65 by FBXW2 suppresses breast cancer stemness, tumorigenesis, and paclitaxel resistance. Cell Death Differ. 2022;29:381–92.
Yin X, Teng X, Ma T, Yang T, Zhang J, Huo M, et al. RUNX2 recruits the NuRD(MTA1)/CRL4B complex to promote breast cancer progression and bone metastasis. Cell Death Differ. 2022;29:2203–17.
Zhao Y, Huang X, Zhu D, Wei M, Luo J, Yu S, et al. Deubiquitinase OTUD6A promotes breast cancer progression by increasing TopBP1 stability and rendering tumor cells resistant to DNA-damaging therapy. Cell Death Differ. 2022;29:2531–44.
Panatta E, Butera A, Celardo I, Leist M, Melino G, Amelio I. p53 regulates expression of nuclear envelope components in cancer cells. Biol Direct. 2022;17:38.
Panatta E, Zampieri C, Melino G, Amelio I. Understanding p53 tumour suppressor network. Biol Direct. 2021;16:14.
Rozenberg JM, Zvereva S, Dalina A, Blatov I, Zubarev I, Luppov D, et al. The p53 family member p73 in the regulation of cell stress response. Biol Direct. 2021;16:23.
Butera A, Roy M, Zampieri C, Mammarella E, Panatta E, Melino G, et al. p53-driven lipidome influences non-cell-autonomous lysophospholipids in pancreatic cancer. Biol Direct. 2022;17:6.
Smirnov A, Lena AM, Cappello A, Panatta E, Anemona L, Bischetti S, et al. ZNF185 is a p63 target gene critical for epidermal differentiation and squamous cell carcinoma development. Oncogene 2019;38:1625–38.
Kudo M, Zalles N, Distefano R, Nigita G, Veneziano D, Gasparini P, et al. Synergistic apoptotic effect of miR-183-5p and Polo-Like kinase 1 inhibitor NMS-P937 in breast cancer cells. Cell Death Differ. 2022;29:407–19.
Jiang D, Qiu T, Peng J, Li S, Tala, Ren W, et al. YB-1 is a positive regulator of KLF5 transcription factor in basal-like breast cancer. Cell Death Differ. 2022;29:1283–95.
Qian J, Lei X, Sun Y, Zheng L, Li J, Zhang S, et al. Long non-coding RNA SNHG8 enhances triple-negative breast cancer cell proliferation and migration by regulating the miR-335-5p/PYGO2 axis. Biol Direct. 2021;16:13.
Liu YY, Liu HY, Yu TJ, Lu Q, Zhang FL, Liu GY, et al. O-GlcNAcylation of MORC2 at threonine 556 by OGT couples TGF-β signaling to breast cancer progression. Cell Death Differ. 2022;29:861–73.
Yan Y, He M, Zhao L, Wu H, Zhao Y, Han L, et al. A novel HIF-2α targeted inhibitor suppresses hypoxia-induced breast cancer stemness via SOD2-mtROS-PDI/GPR78-UPR(ER) axis. Cell Death Differ. 2022;29:1769–89.
**e W, Jiang Q, Wu X, Wang L, Gao B, Sun Z, et al. IKBKE phosphorylates and stabilizes Snail to promote breast cancer invasion and metastasis. Cell Death Differ. 2022;29:1528–40.
Han X, Ren C, Lu C, Qiao P, Yang T, Yu Z. Deubiquitination of MYC by OTUB1 contributes to HK2 mediated glycolysis and breast tumorigenesis. Cell Death Differ. 2022;29:1864–73.
Memmi EM, Sanarico AG, Giacobbe A, Peschiaroli A, Frezza V, Cicalese A, et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2015;112:3499–504.
Cassandri M, Butera A, Amelio I, Lena AM, Montanaro M, Mauriello A, et al. ZNF750 represses breast cancer invasion via epigenetic control of prometastatic genes. Oncogene. 2020;39:4331–43.
Melino G, Memmi EM, Pelicci PG, Bernassola F. Maintaining epithelial stemness with p63. Sci Signal. 2015;8:re9.
Vanni G, Pellicciaro M, Materazzo M, Dauri M, D’Angelillo RM, Buonomo C, et al. Awake breast cancer surgery: strategy in the beginning of COVID-19 emergency. Breast Cancer. 2021;28:137–44.
Butera A, Cassandri M, Rugolo F, Agostini M, Melino G. The ZNF750-RAC1 axis as potential prognostic factor for breast cancer. Cell Death Discov. 2020;6:135.
Zhang J, Zhang G, Zhang W, Bai L, Wang L, Li T, et al. Loss of RBMS1 promotes anti-tumor immunity through enabling PD-L1 checkpoint blockade in triple-negative breast cancer. Cell Death Differ. 2022;29:2247–61.
Sharma P, Klemp JR, Kimler BF, Mahnken JD, Geier LJ, Khan QJ, et al. Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res Treat. 2014;145:707–14.
Wong-Brown MW, Meldrum CJ, Carpenter JE, Clarke CL, Narod SA, Jakubowska A, et al. Prevalence of BRCA1 and BRCA2 germline mutations in patients with triple-negative breast cancer. Breast Cancer Res Treat. 2015;150:71–80.
Borg A, Haile RW, Malone KE, Capanu M, Diep A, Törngren T, et al. Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: the WECARE study. Hum Mutat. 2010;31:E1200–40.
Claes K, Poppe B, Coene I, Paepe AD, Messiaen L. BRCA1 and BRCA2 germline mutation spectrum and frequencies in Belgian breast/ovarian cancer families. Br J Cancer. 2004;90:1244–51.
Scully R, Chen J, Plug A, **ao Y, Weaver D, Feunteun J, et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–75.
Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–32.
Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res. 2001;61:4092–7.
Vitale I, Pietrocola F, Guilbaud E, Aaronson SA, Abrams JM, Adam D, et al. Apoptotic cell death in disease-current understanding of the NCCD 2023. Cell Death Differ. 2023;30:1097–154.
Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25.
Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, et al. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998;16:1713–21.
Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31:860–7.
Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ, et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst. 1998;90:1138–45.
Nolan E, Savas P, Policheni AN, Darcy PK, Vaillant F, Mintoff CP, et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med. 2017;9:eaal4922.
Chabanon RM, Pedrero M, Lefebvre C, Marabelle A, Soria JC, Postel-Vinay S. Mutational landscape and sensitivity to immune checkpoint blockers. Clin Cancer Res. 2016;22:4309–21.
Wimberly H, Brown JR, Schalper K, Haack H, Silver MR, Nixon C, et al. PD-L1 expression correlates with tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy in breast cancer. Cancer Immunol Res. 2015;3:326–32.
Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2:361–70.
Sabatier R, Finetti P, Mamessier E, Adelaide J, Chaffanet M, Ali HR, et al. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget. 2015;6:5449–64.
Wen WX, Leong CO. Association of BRCA1- and BRCA2-deficiency with mutation burden, expression of PD-L1/PD-1, immune infiltrates, and T cell-inflamed signature in breast cancer. PLoS ONE. 2019;14:e0215381.
Gallo M, Paludi D, Cicero DO, Chiovitti K, Millo E, Salis A, et al. Identification of a conserved N-cap** box important for the structural autonomy of the prion alpha 3-helix: the disease associated D202N mutation destabilizes the helical conformation. Int J Immunopathol Pharm. 2005;18:95–112.
Melino S, Nepravishta R, Bellomaria A, Di Marco S, Paci M. Nucleic acid binding of the RTN1-C C-terminal region: toward the functional role of a reticulon protein. Biochemistry. 2009;48:242–53.
Favaloro B, Tamburro A, Angelucci S, Luca AD, Melino S, di Ilio C, et al. Molecular cloning, expression and site-directed mutagenesis of glutathione S-transferase from Ochrobactrum anthropi. Biochem J. 1998;335:573–9. (Pt 3)(Pt 3)
Angelucci S, Sacchetta P, Moio P, Melino S, Petruzzelli R, Gervasi P, et al. Purification and characterization of glutathione transferases from the sea bass (Dicentrarchus labrax) liver. Arch Biochem Biophys. 2000;373:435–41.
Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–83.
Wang A, Papneja A, Hyrcza M, Al-Habeeb A, Ghazarian D. Gene of the month: BAP1. J Clin Pathol. 2016;69:750–3.
Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014;111:285–90.
Laitman Y, Newberg J, Molho RB, ** DX, Friedman E. The spectrum of tumors harboring BAP1 gene alterations. Cancer Genet. 2021;256-257:31–5.
Heeke AL, **u J, Elliott A, Korn WM, Lynce F, Pohlmann PR, et al. Actionable co-alterations in breast tumors with pathogenic mutations in the homologous recombination DNA damage repair pathway. Breast Cancer Res Treat. 2020;184:265–75.
Venkitaraman AR. Tumour suppressor mechanisms in the control of chromosome stability: insights from BRCA2. Mol Cells. 2014;37:95–9.
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X, et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35:428–40.e5.
Gao Y, Zhang W, Liu C, Li G. miR-200 affects tamoxifen resistance in breast cancer cells through regulation of MYB. Sci Rep. 2019;9:18844.
Sakura H, Kanei-Ishii C, Nagase T, Nakagoshi H, Gonda TJ, Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci USA. 1989;86:5758–62.
Clifford N, Smith LM, Powell J, Gattenlöhner S, Marx A, O’Connor R. The EphA3 receptor is expressed in a subset of rhabdomyosarcoma cell lines and suppresses cell adhesion and migration. J Cell Biochem. 2008;105:1250–9.
Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74.
Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75.
Janes PW, Vail ME, Gan HK, Scott AM. Antibody targeting of Eph receptors in cancer. Pharmaceuticals. 2020;13:88.
Swords RT, Greenberg PL, Wei AH, Durrant S, Advani AS, Hertzberg MS, et al. KB004, a first in class monoclonal antibody targeting the receptor tyrosine kinase EphA3, in patients with advanced hematologic malignancies: results from a phase 1 study. Leuk Res. 2016;50:123–31.
Itoh T, Hatano R, Horimoto Y, Yamada T, Song D, Otsuka H, et al. IL-26 mediates epidermal growth factor receptor-tyrosine kinase inhibitor resistance through endoplasmic reticulum stress signaling pathway in triple-negative breast cancer cells. Cell Death Dis. 2021;12:520.
Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949.
Lisabeth EM, Fernandez C, Pasquale EB. Cancer somatic mutations disrupt functions of the EphA3 receptor tyrosine kinase through multiple mechanisms. Biochemistry. 2012;51:1464–75.
Bonfiglio R, Nardozi D, Scimeca M, Cerroni C, Mauriello A, Bonanno E. PD-L1 in immune-escape of breast and prostate cancers: from biology to therapy. Future Oncol. 2017;13:2129–31.
Scimeca M, Bonfiglio R, Urbano N, Cerroni C, Anemona L, Montanaro M, et al. Programmed death ligand 1 expression in prostate cancer cells is associated with deep changes of the tumor inflammatory infiltrate composition. Urol Oncol. 2019;37:297.e19–e31.
Cerniglia M, **u J, Grothey A, Pishvaian MJ, Baca Y, Hwang JJ, et al. Association of homologous recombination-DNA damage response gene mutations with immune biomarkers in gastroesophageal cancers. Mol Cancer Ther. 2022;21:227–36.
Corvigno S, Burks JK, Hu W, Zhong Y, Jennings NB, Fleming ND, et al. Immune microenvironment composition in high-grade serous ovarian cancers based on BRCA mutational status. J Cancer Res Clin Oncol. 2021;147:3545–55.
Domchek SM, Postel-Vinay S, Im SA, Park YH, Delord JP, Italiano A, et al. Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): an open-label, multicentre, phase 1/2, basket study. Lancet Oncol. 2020;21:1155–64.
Vinayak S, Tolaney SM, Schwartzberg L, Mita M, McCann G, Tan AR, et al. Open-label Clinical Trial of Niraparib Combined With Pembrolizumab for Treatment of Advanced or Metastatic Triple-Negative Breast Cancer. JAMA Oncol. 2019;5:1132–40.
Gezgin G, Dogrusöz M, van Essen TH, Kroes WGM, Luyten GPM, van der Velden PA, et al. Genetic evolution of uveal melanoma guides the development of an inflammatory microenvironment. Cancer Immunol Immunother. 2017;66:903–12.
Shrestha R, Nabavi N, Lin YY, Mo F, Anderson S, Volik S, et al. BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. Genome Med. 2019;11:8.
Wang T, Lu R, Kapur P, Jaiswal BS, Hannan R, Zhang Z, et al. An empirical approach leveraging tumorgrafts to dissect the tumor microenvironment in renal cell carcinoma identifies missing link to prognostic inflammatory factors. Cancer Discov. 2018;8:1142–55.
Figueiredo CR, Kalirai H, Sacco JJ, Azevedo RA, Duckworth A, Slupsky JR, et al. Loss of BAP1 expression is associated with an immunosuppressive microenvironment in uveal melanoma, with implications for immunotherapy development. J Pathol. 2020;250:420–39.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Martorana F, Motta G, Pavone G, Motta L, Stella S, Vitale SR, et al. AKT inhibitors: new weapons in the fight against breast cancer? Front Pharm. 2021;12:662232.
Shi Z, Wulfkuhle J, Nowicka M, Gallagher RI, Saura C, Nuciforo PG, et al. Functional map** of AKT signaling and biomarkers of response from the FAIRLANE trial of neoadjuvant ipatasertib plus paclitaxel for triple-negative breast cancer. Clin Cancer Res. 2022;28:993–1003.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Van der Auwera GA, O’Connor BD. Genomics in the cloud: using Docker, GATK, and WDL in Terra: O’Reilly Media; 2020.
Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Källberg M, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–4.
Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–5.
Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–7.
Ha G, Roth A, Khattra J, Ho J, Yap D, Prentice LM, et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 2014;24:1881–93.
Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i9.
Bekker-Jensen DB, Bernhardt OM, Hogrebe A, Martinez-Val A, Verbeke L, Gandhi T, et al. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat Commun. 2020;11:787.
Bruderer R, Muntel J, Müller S, Bernhardt OM, Gandhi T, Cominetti O, et al. Analysis of 1508 plasma samples by capillary-flow data-independent acquisition profiles proteomics of weight loss and maintenance. Mol Cell Proteom. 2019;18:1242–54.
Manders F, Brandsma AM, de Kanter J, Verheul M, Oka R, van Roosmalen MJ, et al. MutationalPatterns: the one stop shop for the analysis of mutational processes. BMC Genomics. 2022;23:134.
Huang MN, McPherson JR, Cutcutache I, Teh BT, Tan P, Rozen SG. MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci Rep. 2015;5:13321.
Gendoo DM, Ratanasirigulchai N, Schröder MS, Paré L, Parker JS, Prat A, et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics. 2016;32:1097–9.
Funding
The Research leading to these results has received funding from the MUR-PNRR M4C2I1.3 PE6 project PE00000019 Heal Italia (CUP: E83C22004670001) to GM, EC, and FB. The Research leading to these results has received funding from AIRC under IG 2022 - ID. 27366 project – P.I. Melino Gennaro; under IG 2019 - ID. 22206 project – P.I. Candi Eleonora; under IG 2019 – 23232 project – P.I. Bernassola Francesca. Fondazione Luigi Maria Monti IDI-IRCCS (R.C. to EC). Work has been also supported by Regione Lazio through LazioInnova Progetto Gruppo di Ricerca n 85-2017-14986; n 33 & 55-2021-T0002E0001.
Author information
Authors and Affiliations
Consortia
Contributions
GM, EC, and FB conceived the project; YH, GM, YS, AS, JB, and FB wrote the manuscript; YH, AS, and JB prepared figures. All authors have approved this submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Compliance with Ethical Norms
All the procedures carried out in the research with participation of humans were in compliance with the ethical standards of the institutional and/or national ethics committee and with the Helsinki Declaration of 1964 and its subsequent changes or with comparable ethics standards. Informed voluntary consent was obtained from every participant of the study: approval on 09-2019, number 96-19.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, Y., Rovella, V., Smirnov, A. et al. A BRCA2 germline mutation and high expression of immune checkpoints in a TNBC patient. Cell Death Discov. 9, 370 (2023). https://doi.org/10.1038/s41420-023-01651-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-023-01651-3
- Springer Nature Limited
This article is cited by
-
Molecular profiling of a bladder cancer with very high tumour mutational burden
Cell Death Discovery (2024)
-
Genetically driven predisposition leads to an unusually genomic unstable renal cell carcinoma
Discover Oncology (2024)