Abstract
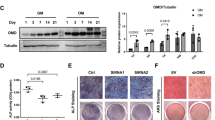
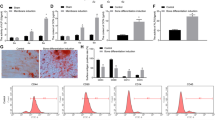
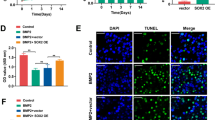
TNF receptor-associated factor 4 (TRAF4), a member of the TRAF family, plays an important role in the embryogenesis and development of the bone system. Mesenchymal stem cells (MSCs), which are the primary origin of osteoblasts in vivo, are key cells in bone development; however, whether TRAF4 modulates the osteogenic capacity of MSCs has never been explored. In this study, we demonstrated that TRAF4 positively regulates the osteogenic process of MSCs both in vitro and in vivo. In addition, we further demonstrated that TRAF4 modulates the osteogenic process of MSCs by acting as an E3 ubiquitin ligase to mediate the K48-linked ubiquitination of Smurf2 at the K119 site and cause degradation. Furthermore, TRAF4 was abnormally decreased in bone sections of ovariectomized rat and osteoporosis patients. Taken together, our findings suggest that TRAF4 positively regulates the osteogenic differentiation of MSCs by acting as an E3 ubiquitin ligase to degrade Smurf2. These results emphasize the critical role of TRAF4 in bone formation and could not only improve the clinical use of MSCs in tissue engineering but also clarify the pathogenesis of bone metabolism disorders.
Similar content being viewed by others
Introduction
Mesenchymal stem cells (MSCs) are multipotent progenitor cells that can differentiate into osteoblasts, chondrocytes, and adipocytes [1, 2]. Due to their strong potential in osteogenic differentiation capacity, MSCs are considered to be the most promising cell types used in tissue engineering technology for bone regeneration and repair [3,4,5]. However, the molecular mechanism that governs the osteogenic differentiation of MSCs remains largely unknown and hampers the further development of MSC-based cell therapies for bone defects in the clinic. Therefore, to efficiently harness MSCs for therapeutic purposes, it is necessary to understand the molecular mechanisms underlying MSC osteogenic differentiation.
TNF receptor-associated factor 4 (TRAF4) is a unique member of the TRAF family [6]. Previous in vivo studies have clearly demonstrated that TRAF4 is involved in embryogenesis and that TRAF4 deficiency results in severe malformation of the skeleton system, which indicates that TRAF4 is indispensable in bone development [7]. However, further studies illustrating the concrete role of TRAF4 in the process of bone remodeling at the molecular biological level have not been reported. Osteoblasts are among the most important cells in regulating the bone remodeling process in vivo [8, 9], and MSCs are the major origin of osteoblasts [10, 11]. However, whether TRAF4 modulates the osteogenic capacity of MSCs has never been explored.
Smad ubiquitination-related factor 2 (Smurf2) is a member of the Hect domain family of E3 ubiquitin ligases [12, 13], and it interacts with and degrades various essential osteogenesis-related molecules including Smad1 and Runx2 to negatively regulate the osteogenic differentiation process [12, 14, 15]. Although the mechanism of Smurf2 in modulating the osteogenic differentiation process has been widely studied, the upper regulatory network to modulate the expression of Smurf2 during the osteogenic differentiation process of MSCs remains unclear.
Taken together, our findings suggest that TRAF4 positively regulates the osteogenic process of MSCs both in vitro and in vivo, can be a promising target not only to improve the efficiency of MSC-based tissue engineering therapy for bone regeneration and repair but also to treat the pathogenesis of bone metabolism disorders, such as osteoporosis.
Methods and materials
Isolation and culture of MSCs
This study was approved by the ethics committee of Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, China. Written informed consent was obtained from all subjects included in the study. MSCs were isolated and cultured as previously described [ Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. Watson L, Elliman SJ, Coleman CM. From isolation to implantation: a concise review of mesenchymal stem cell therapy in bone fracture repair. Stem Cell Res Ther. 2014;5:51. Jones E, Yang X. Mesenchymal stem cells and bone regeneration: current status. Injury. 2011;42:562–568. Garg P, Mazur MM, Buck AC, Wandtke ME, Liu J, Ebraheim NA. Prospective review of mesenchymal stem cells differentiation into osteoblasts. Orthop Surg. 2017;9:13–19. Kedinger V, Rio MC. TRAF4, the unique family member. Adv Exp Med Biol. 2007;597:60–71. Regnier CH, Masson R, Kedinger V, Textoris J, Stoll I, Chenard MP, et al. Impaired neural tube closure, axial skeleton malformations, and tracheal ring disruption in TRAF4-deficient mice. Proc Natl Acad Sci USA. 2002;99:5585–5590. Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–355. Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8:272–288. Heino TJ, Hentunen TA. Differentiation of osteoblasts and osteocytes from mesenchymal stem cells. Curr Stem Cell Res Ther. 2008;3:131–145. Abdallah BM, Alzahrani AM, Kassem M. Secreted clusterin protein inhibits osteoblast differentiation of bone marrow mesenchymal stem cells by suppressing ERK1/2 signaling pathway. Bone. 2018;110:221–229. Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98:974–979. Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. Kaneki H, Guo R, Chen D, Yao Z, Schwarz EM, Zhang YE, et al. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem. 2006;281:4326–4333. Choi YH, Kim YJ, Jeong HM, ** YH, Yeo CY, Lee KY. Akt enhances Runx2 protein stability by regulating Smurf2 function during osteoblast differentiation. FEBS J. 2014;281:3656–3666. **e Z, Wang P, Li Y, Deng W, Zhang X, Su H, et al. Imbalance between bone morphogenetic protein 2 and noggin induces abnormal osteogenic differentiation of mesenchymal stem cells in ankylosing spondylitis. Arthritis Rheumatol. 2016;68:430–440. Li M, **e Z, Wang P, Li J, Liu W, Tang S, et al. The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis. 2018;9:554. **e Z, Tang SA, Ye G, Wang P, Li J, Liu W, et al. Interleukin-6/interleukin-6 receptor complex promotes osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Stem Cell Res Ther. 2018;9:13. Gregory CA, Gunn WG, Peister A, Prockop DJ. An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem. 2004;329:77–84. Wu LN, Ishikawa Y, Sauer GR, Genge BR, Mwale F, Mishima H, et al. Morphological and biochemical characterization of mineralizing primary cultures of avian growth plate chondrocytes: evidence for cellular processing of Ca2+ and Pi prior to matrix mineralization. J Cell Biochem. 1995;57:218–237. Vaisman DN, McCarthy AD, Cortizo AM. Bone-specific alkaline phosphatase activity is inhibited by bisphosphonates: role of divalent cations. Biol Trace Elem Res. 2005;104:131–140. Sakano S, Murata Y, Miura T, Iwata H, Sato K, Matsui N, et al. Collagen and alkaline phosphatase gene expression during bone morphogenetic protein (BMP)-induced cartilage and bone differentiation. Clin Orthop Relat Res. 1993:337–344. https://doi.org/10.1097/00003086-199307000-00044. Kum CJ, Kim EY, Kim JH, Lee B, Min JH, Heo J, et al. Cyperus Rotundus L. extract suppresses RANKL-induced osteoclastogenesis through NFATc1/c-fos downregulation and prevent bone loss in OVX-induced osteoporosis rat. J Ethnopharmacol. 2017;205:186–194. Zhang P, Liu Y, ** C, Zhang M, Lv L, Zhang X, et al. Histone H3K9 Acetyltransferase PCAF is essential for osteogenic differentiation through bone morphogenetic protein signaling and may be involved in osteoporosis. Stem Cells. 2016;34:2332–2341. Singh R, Karri D, Shen H, Shao J, Dasgupta S, Huang S, et al. TRAF4-mediated ubiquitination of NGF receptor TrkA regulates prostate cancer metastasis. J Clin Invest. 2018;128:3129–3143. Li W, Peng C, Lee MH, Lim D, Zhu F, Fu Y, et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 2013;73:6938–6950. Zhang L, Zhou F, de Vinuesa AG, de Kruijf EM, Mesker WE, Hui L, et al. TRAF4 promotes TGF-beta receptor signaling and drives breast cancer metastasis. Mol Cell. 2013;51:559–572. Zepp JA, Wu L, Qian W, Ouyang W, Aronica M, Erzurum S, et al. TRAF4-SMURF2-mediated DAZAP2 degradation is critical for IL-25 signaling and allergic airway inflammation. J Immunol. 2015;194:2826–2837. Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;423:995–999. Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. Black DM, Rosen CJ. Clinical practice. Postmenopausal osteoporosis. N Engl J Med. 2016;374:254–262. Ardawi MM, Badawoud MH, Hassan SM, Rouzi AA, Ardawi JMS, AlNosani NM, et al. Lycopene treatment against loss of bone mass, microarchitecture and strength in relation to regulatory mechanisms in a postmenopausal osteoporosis model. Bone. 2016;83:127–140. Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13:27–38. Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. Chang J, Liu F, Lee M, Wu B, Ting K, Zara JN, et al. NF-kappaB inhibits osteogenic differentiation of mesenchymal stem cells by promoting beta-catenin degradation. Proc Natl Acad Sci USA. 2013;110:9469–9474. Kumar Y, Kapoor I, Khan K, Thacker G, Khan MP, Shukla N, et al. E3 ubiquitin ligase Fbw7 negatively regulates osteoblast differentiation by targeting Runx2 for degradation. J Biol Chem. 2015;290:30975–30987. Li X, Huang M, Zheng H, Wang Y, Ren F, Shang Y, et al. CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J Cell Biol. 2008;181:959–972. Choi HK, Yuan H, Fang F, Wei X, Liu L, Li Q, et al. Tsc1 regulates the balance between osteoblast and adipocyte differentiation through autophagy/notch1/beta-catenin cascade. J Bone Miner Res. 2018;33:2021–2034. Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4:85–87. McKeon JE, Sha D, Li L, Chin LS. Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell Mol Life Sci. 2015;72:1811–1824. Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–126. Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–1253. Chen YY, Rosario FJ, Shehab MA, Powell TL, Gupta MB, Jansson T. Increased ubiquitination and reduced plasma membrane trafficking of placental amino acid transporter SNAT-2 in human IUGR. Clin Sci. 2015;129:1131–1141.References
Acknowledgements
This study was financially supported by grants from the National Natural Science Foundation of China (81672097 and 31700884) and the Science and Technology Project of Guangdong Province (2015B020228001).
Author information
Authors and Affiliations
Contributions
JL, PW, ZX, SW, and SC performed the experiments. ML, WL, and ST collected the human samples. HS, MM, and XW isolated and cultured the MSCs. JL, ZX, GY, and GZ wrote the initial draft of the paper. HS, YW, PW, JL, and ZX contributed to the conceptualization and experimental design of the study and edited the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by D. Aberdam
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Wang, P., **e, Z. et al. TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ 26, 2652–2666 (2019). https://doi.org/10.1038/s41418-019-0328-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0328-3
- Springer Nature Limited
This article is cited by
-
Mechanical force induces macrophage-derived exosomal UCHL3 promoting bone marrow mesenchymal stem cell osteogenesis by targeting SMAD1
Journal of Nanobiotechnology (2023)
-
TRAF4 hyperactivates HER2 signaling and contributes to Trastuzumab resistance in HER2-positive breast cancer
Oncogene (2022)
-
The N6-methyladenosine demethylase ALKBH5 negatively regulates the osteogenic differentiation of mesenchymal stem cells through PRMT6
Cell Death & Disease (2021)
-
Structural feature of TRAFs, their related human diseases and therapeutic intervention
Archives of Pharmacal Research (2021)