Abstract
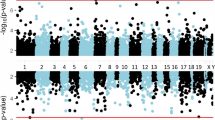
Epigenetic factors modify the effects of environmental factors on biological outcomes. Identification of epigenetic changes that associate with PTSD is therefore a crucial step in deciphering mechanisms of risk and resilience. In this study, our goal is to identify epigenetic signatures associated with PTSD symptom severity (PTSS) and changes in PTSS over time, using whole blood DNA methylation (DNAm) data (MethylationEPIC BeadChip) of military personnel prior to and following combat deployment. A total of 429 subjects (858 samples across 2 time points) from three male military cohorts were included in the analyses. We conducted two different meta-analyses to answer two different scientific questions: one to identify a DNAm profile of PTSS using a random effects model including both time points for each subject, and the other to identify a DNAm profile of change in PTSS conditioned on pre-deployment DNAm. Four CpGs near four genes (F2R, CNPY2, BAIAP2L1, and TBXAS1) and 88 differentially methylated regions (DMRs) were associated with PTSS. Change in PTSS after deployment was associated with 15 DMRs, of those 2 DMRs near OTUD5 and ELF4 were also associated with PTSS. Notably, three PTSS-associated CpGs near F2R, BAIAP2L1 and TBXAS1 also showed nominal evidence of association with change in PTSS. This study, which identifies PTSD-associated changes in genes involved in oxidative stress and immune system, provides novel evidence that epigenetic differences are associated with PTSS.

Similar content being viewed by others
Data availability
The main summary statistics data that support the findings of this study are available within Supplementary Data. Owing to military cohort data sharing restrictions, data from MRS, Army STARRS, and PRISMO cannot be publicly posted. Individual-level data from the cohorts or cohort-level summary statistics will be made available to researchers following an approved analysis proposal through the PGC Post-traumatic Stress Disorder group with agreement of the cohort PIs. For additional information on access to these data, including PI contact information for the contributing cohorts, please contact the corresponding author.
Code availability
The scripts generated to perform the Meta-Analysis 1, Meta-Analysis 2, and DMR analysis are available in https://github.com/PGC-PTSD-EWAS/PGC-PTSD-Longitudinal-Analysis.
References
American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-5®). 5th ed. Washington, DC: American Psychiatric Pub; 2013.
Benjet C, Bromet E, Karam EG, Kessler RC, McLaughlin KA, Ruscio AM, et al. The epidemiology of traumatic event exposure worldwide: results from the World Mental Health Survey Consortium. Psychol Med. 2016;46:327–43.
Kessler RC, Berglund P, Demler O, ** R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602.
Daskalakis NP, Rijal CM, King C, Huckins LM, Ressler KJ. Recent genetics and epigenetics approaches to PTSD. Curr Psychiatry Rep. 2018;20:30.
Morrison FG, Miller MW, Logue MW, Assef M, Wolf EJ. DNA methylation correlates of PTSD: recent findings and technical challenges. Prog Neuropsychopharmacol Biol Psychiatry. 2019;90:223–34.
Sheerin CM, Lind MJ, Bountress KE, Nugent NR, Amstadter AB. The genetics and epigenetics of PTSD: overview, recent advances, and future directions. Curr Opin Psychol. 2017;14:5–11.
Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci USA. 2010;107:9470–5.
Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, et al. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:700–8.
Kuan PF, Waszczuk MA, Kotov R, Marsit CJ, Guffanti G, Gonzalez A, et al. An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Transl Psychiatry. 2017;7:e1158.
Uddin M, Ratanatharathorn A, Armstrong D, Kuan PF, Aiello AE, Bromet EJ, et al. Epigenetic meta-analysis across three civilian cohorts identifies NRG1 and HGS as blood-based biomarkers for post-traumatic stress disorder. Epigenomics. 2018;10:1585–601.
Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, et al. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry. 2018;23:1145–56.
Snijders C, Maihofer AX, Ratanatharathorn A, Baker DG, Boks MP, Geuze E, et al. Longitudinal epigenome-wide association studies of three male military cohorts reveal multiple CpG sites associated with post-traumatic stress disorder. Clin Epigenetics. 2020;12:11.
Logue MW, Miller MW, Wolf EJ, Huber BR, Morrison FG, Zhou Z, et al. An epigenome-wide association study of posttraumatic stress disorder in US veterans implicates several new DNA methylation loci. Clin Epigenetics. 2020;12:46.
Smith AK, Ratanatharathorn A, Maihofer AX, Naviaux RK, Aiello AE, Amstadter AB, et al. Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies methylation changes in AHRR. Nat Commun. 2020;11:5965.
Verhulst B, Neale MC. Best practices for binary and ordinal data analyses. Behav Genet. 2021;51:204–14.
Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28:1280–1.
McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom Data. 2016;9:22–4.
Fortin JP, Triche TJ Jr., Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–60.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3.
Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11:587.
Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinform. 2017;18:105.
Salas LA, Koestler DC, Butler RA, Hansen HM, Wiencke JK, Kelsey KT, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19:64.
Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R, et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38:231–41.
Ratanatharathorn A, Boks MP, Maihofer AX, Aiello AE, Amstadter AB, Ashley-Koch AE, et al. Epigenome-wide association of PTSD from heterogeneous cohorts with a common multi-site analysis pipeline. Am J Med Genet B Neuropsychiatr Genet. 2017;174:619–30.
de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–8.
Lee CH, Cook S, Lee JS, Han B. Comparison of two meta-analysis methods: inverse-variance-weighted average and weighted sum of Z-scores. Genomics Inform. 2016;14:173–80.
Cochran WG. Some methods for strengthening the common χ2 tests. Biometrics. 1954;10:417–51.
Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genom. 2019;20:366.
Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, Reginald VL, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6
Braun PR, Han S, Hing B, Nagahama Y, Gaul LN, Heinzman JT, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiatry. 2019;9:47.
Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–8.
Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32:286–8.
Nemeroff CB, Bremner JD, Foa EB, Mayberg HS, North CS, Stein MB. Posttraumatic stress disorder: a state-of-the-science review. J Psychiatr Res. 2006;40:1–21.
Yehuda R. Risk and resilience in posttraumatic stress disorder. J Clin Psychiatry. 2004;65:29–36.
Brewin CR, Holmes EA. Psychological theories of posttraumatic stress disorder. Clin Psychol Rev. 2003;23:339–76.
Jose RJ, Williams AE, Chambers RC. Proteinase-activated receptors in fibroproliferative lung disease. Thorax. 2014;69:190–2.
Tylee DS, Chandler SD, Nievergelt CM, Liu X, Pazol J, Woelk CH, et al. Blood-based gene-expression biomarkers of post-traumatic stress disorder among deployed marines: a pilot study. Psychoneuroendocrinology. 2015;51:472–94.
Hong F, Liu B, Wu BX, Morreall J, Roth B, Davies C, et al. CNPY2 is a key initiator of the PERK-CHOP pathway of the unfolded protein response. Nat Struct Mol Biol. 2017;24:834–9.
Bornhauser BC, Olsson PA, Lindholm D. MSAP is a novel MIR-interacting protein that enhances neurite outgrowth and increases myosin regulatory light chain. J Biol Chem. 2003;278:35412–20.
Miller MW, Lin AP, Wolf EJ, Miller DR. Oxidative stress, inflammation, and neuroprogression in chronic PTSD. Harv Rev Psychiatry. 2018;26:57–69.
Wang YP, Huang LY, Sun WM, Zhang ZZ, Fang JZ, Wei BF, et al. Insulin receptor tyrosine kinase substrate activates EGFR/ERK signalling pathway and promotes cell proliferation of hepatocellular carcinoma. Cancer Lett. 2013;337:96–106.
Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007;120:1663–72.
Alhassen S, Chen S, Alhassen L, Phan A, Khoudari M, De Silva A, et al. Intergenerational stress transmission is associated with brain metabotranscriptome remodeling and mitochondrial dysfunction. Commun Biol. 2021;4:783.
Woodward DF, Jones RL, Narumiya S. International Union of Basic and Clinical Pharmacology. LXXXIII: classification of prostanoid receptors, updating 15 years of progress. Pharmacol Rev. 2011;63:471–538.
Offermanns S. Activation of platelet function through G protein-coupled receptors. Circ Res. 2006;99:1293–304.
Smyth EM. Thromboxane and the thromboxane receptor in cardiovascular disease. Clin Lipido. 2010;5:209–19.
Sadowitz PD, Setty BN, Stuart M. The platelet cyclooxygenase metabolite 12-L-hydroxy-5, 8, 10-hepta-decatrienoic acid (HHT) may modulate primary hemostasis by stimulating prostacyclin production. Prostaglandins. 1987;34:749–63.
Campbell PB, Tolson TA. Modulation of human monocyte leukotactic responsiveness by thromboxane A2 and 12-hydroxyheptadecatrienoic acid (12-HHT). J Leukoc Biol. 1988;43:117–24.
Hecker M, Ullrich V. 12(S)-Hydroxy-5,8,10 (Z,E,E)-heptadecatrienoic acid (HHT) is preferentially metabolized to its 12-keto derivative by human erythrocytes in vitro. Eicosanoids. 1988;1:19–25.
Del Rio D, Stewart AJ, Pellegrini N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr Metab Cardiovasc Dis. 2005;15:316–28.
Xue SS, He JL, Zhang X, Liu YJ, Xue FX, Wang CJ, et al. Metabolomic analysis revealed the role of DNA methylation in the balance of arachidonic acid metabolism and endothelial activation. Biochim Biophys Acta. 2015;1851:1317–26.
Narumiya S, Sugimoto Y, Ushikubi F. Prostanoid receptors: structures, properties, and functions. Physiol Rev. 1999;79:1193–226.
Mitsumori T, Furuyashiki T, Momiyama T, Nishi A, Shuto T, Hayakawa T, et al. Thromboxane receptor activation enhances striatal dopamine release, leading to suppression of GABAergic transmission and enhanced sugar intake. Eur J Neurosci. 2011;34:594–604.
Dahoun T, Nour MM, McCutcheon RA, Adams RA, Bloomfield MAP, Howes OD. The relationship between childhood trauma, dopamine release and dexamphetamine-induced positive psychotic symptoms: a [(11)C]-(+)-PHNO PET study. Transl Psychiatry. 2019;9:287.
Bloomfield MA, McCutcheon RA, Kempton M, Freeman TP, Howes O. The effects of psychosocial stress on dopaminergic function and the acute stress response. Elife. 2019;8:e46797.
Howes OD, Montgomery AJ, Asselin MC, Murray RM, Valli I, Tabraham P, et al. Elevated striatal dopamine function linked to prodromal signs of schizophrenia. Arch Gen Psychiatry. 2009;66:13–20.
Avram M, Brandl F, Cabello J, Leucht C, Scherr M, Mustafa M, et al. Reduced striatal dopamine synthesis capacity in patients with schizophrenia during remission of positive symptoms. Brain. 2019;142:1813–26.
Boukezzi S, Baunez C, Rousseau PF, Warrot D, Silva C, Guyon V, et al. Posttraumatic Stress Disorder is associated with altered reward mechanisms during the anticipation and the outcome of monetary incentive cues. Neuroimage Clin. 2020;25:102073.
Wang Q, **ang B, Deng W, Wu J, Li M, Ma X, et al. Genome-wide association analysis with gray matter volume as a quantitative phenotype in first-episode treatment-naive patients with schizophrenia. PLoS ONE. 2013;8:e75083.
Maihofer AX, Choi KW, Coleman JRI, Daskalakis NP, Denckla CA, Ketema E, et al. Enhancing discovery of genetic variants for PTSD through integration of quantitative phenotypes and trauma exposure information. Biological Psychiatry. 2021. e-pub ahead of print. https://doi.org/10.1016/j.biopsych.2021.09.020.
Lindsay S, Monk M, Holliday R, Huschtscha L, Davies KE, Riggs AD, et al. Differences in methylation on the active and inactive human X chromosomes. Ann Hum Genet. 1985;49:115–27.
Yu S, Chen C, Pan Y, Kurz MC, Datner E, Hendry PL, et al. Genes known to escape X chromosome inactivation predict co-morbid chronic musculoskeletal pain and posttraumatic stress symptom development in women following trauma exposure. Am J Med Genet B Neuropsychiatr Genet. 2019;180:415–27.
Tanaka M, Li H, Zhang X, Singh J, Dalgard CL, Wilkerson M, et al. Region- and time-dependent gene regulation in the amygdala and anterior cingulate cortex of a PTSD-like mouse model. Mol Brain. 2019;12:25.
Grybek V, Aubry L, Maupetit-Mehouas S, Le Stunff C, Denis C, Girard M, et al. Methylation and transcripts expression at the imprinted GNAS locus in human embryonic and induced pluripotent stem cells and their derivatives. Stem Cell Rep. 2014;3:432–43.
Plongthongkum N, van Eijk KR, de Jong S, Wang T, Sul JH, Boks MP, et al. Characterization of genome-methylome interactions in 22 nuclear pedigrees. PLoS ONE. 2014;9:e99313.
Castellani CA, Laufer BI, Melka MG, Diehl EJ, O’Reilly RL, Singh SM. DNA methylation differences in monozygotic twin pairs discordant for schizophrenia identifies psychosis related genes and networks. BMC Med Genom. 2015;8:17.
Sokolowski M, Wasserman J, Wasserman D. Gene-level associations in suicide attempter families show overrepresentation of synaptic genes and genes differentially expressed in brain development. Am J Med Genet B Neuropsychiatr Genet. 2018;177:774–84.
Muhie S, Gautam A, Chakraborty N, Hoke A, Meyerhoff J, Hammamieh R, et al. Molecular indicators of stress-induced neuroinflammation in a mouse model simulating features of post-traumatic stress disorder. Transl Psychiatry. 2017;7:e1135.
Shu C, Zhang X, Aouizerat BE, Xu K. Comparison of methylation capture sequencing and infinium methylationEPIC array in peripheral blood mononuclear cells. Epigenetics Chromatin. 2020;13:51.
Michopoulos V, Powers A, Gillespie CF, Ressler KJ, Jovanovic T. Inflammation in fear- and anxiety-based disorders: PTSD, GAD, and beyond. Neuropsychopharmacology. 2017;42:254–70.
Acknowledgements
This work was supported by the National Institute of Mental Health (NIMH; 2R01MH108826 and 2R01MH106595). The Marine Corps, Navy Bureau of Medicine and Surgery (BUMED) and VA Health Research and Development (HSR&D) provided funding for MRS data collection. Acknowledged are Mark A. Geyer (UCSD), Daniel T. O’Connor (UCSD), and all MRS investigators, as well as the MRS administrative core and data collection staff. Data collection of PRISMO was funded by the Dutch Ministry of Defense, and DNAm analyses were funded by the VIDI Award fellowship from the Netherlands Organization for Scientific Research (NWO, grant number 917.18.336 to BPFR). Army STARRS was sponsored by the Department of the Army and funded under cooperative agreement number U01MH087981 (2009–2015) with the National Institutes of Health, National Institute of Mental Health (NIH/NIMH).
Author information
Authors and Affiliations
Contributions
Interpreted results, writing, and editing the paper: SK, AKS, and MU. Conceptualization and supervision of project: AKS, MWL, MU, and CMN. Sample and metadata collection: DGB, MPB, EG, RCK, VBR, BPFR, MBS, RJU, EV, MWL, and CMN. Sample preparation: JRP. Formal data analysis: SK, AXM, AW, EK, and AR.
Corresponding author
Ethics declarations
Competing interests
MU was a paid consultant for System Analytic. In the past 3 years, RCK was a consultant for Datastat, Inc., Holmusk, RallyPoint Networks, Inc., and Sage Pharmaceuticals. He has stock options in Mirah, PYM, and Roga Sciences. No other author declares any competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Katrinli, S., Maihofer, A.X., Wani, A.H. et al. Epigenome-wide meta-analysis of PTSD symptom severity in three military cohorts implicates DNA methylation changes in genes involved in immune system and oxidative stress. Mol Psychiatry 27, 1720–1728 (2022). https://doi.org/10.1038/s41380-021-01398-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01398-2
- Springer Nature Limited
This article is cited by
-
Differential methylation of linoleic acid pathway genes is associated with PTSD symptoms – a longitudinal study with Burundian soldiers returning from a war zone
Translational Psychiatry (2024)
-
Prominent genetic variants and epigenetic changes in post-traumatic stress disorder among combat veterans
Molecular Biology Reports (2024)
-
Evaluation of the pooled sample method in Infinium MethylationEPIC BeadChip array by comparison with individual samples
Clinical Epigenetics (2023)
-
DNA methylation GrimAge acceleration in US military veterans with PTSD
Neuropsychopharmacology (2023)
-
Psychological and biological mechanisms linking trauma with cardiovascular disease risk
Translational Psychiatry (2023)