Abstract
Deregulation of epigenetic mechanisms, including microRNA, contributes to leukemogenesis and drug resistance by interfering with cancer-specific molecular pathways. Here, we show that the balance between miR-194-5p and its newly discovered target BCL2-associated transcription factor 1 (BCLAF1) regulates differentiation and survival of normal hematopoietic progenitors. In acute myeloid leukemias this balance is perturbed, locking cells into an immature, potentially ‘immortal’ state. Enhanced expression of miR-194-5p by treatment with the histone deacetylase inhibitor SAHA or by exogenous miR-194-5p expression re-sensitizes cells to differentiation and apoptosis by inducing BCLAF1 to shuttle between nucleus and cytosol. miR-194-5p/BCLAF1 balance was found commonly deregulated in 60 primary acute myeloid leukemia patients and was largely restored by ex vivo SAHA treatment. Our findings link treatment responsiveness to re-instatement of miR-194-5p/BCLAF1 balance.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are endogenous, single-stranded, non-coding RNAs ~18–22 nucleotides long. miRNAs have gene regulatory capability as they are important regulators of post-transcriptional gene expression. miRNAs do not shut down gene expression, but rather fine-tune target’s expression level.1 By targeting multiple genes simultaneously, single (or families of) miRNAs can redirect biological pathways, balancing development, differentiation, apoptosis, stemness and proliferation.1, 2 miRNA functions are cell-, tissue- and disease-specific.3
A global imbalance of miRNA expression (and function) was reported in both solid and hematological malignancies4 including acute myeloid leukemia (AML).5 AML includes genetically diverse malignancies characterized by frequent chromosome translocations and variable response to treatment.6, 7
Promising clinical approaches involve the use of drugs able to modulate deregulated epigenetic processes, such as histone deacetylase inhibitors (HDACi). HDACi were shown to revert the aberrant cancer-associated epigenetic state8, 9 by regulating gene and miRNA expression in several solid tumors10, 11, 12 and hematological cancers.13 It was demonstrated that some HDACi exert beneficial sensitizing effects in current medical treatments although the mechanisms of sensitization are not well understood.14, 15
Here, we identify and characterize the mechanism(s) by which miR-194-5p and its target BCL2-associated transcription factor 1 (BCLAF1) regulate cell cycle progression and differentiation fate. We show that miR-194-5p triggers BCLAF1 downregulation, establishes an euchromatic state typical of differentiating cells, and is associated with improved differentiation ability and response to drugs. We found that miR-194-5p/BCLAF1 balance is deregulated in AML cell lines and ex vivo primary AML blasts, and can at least partially be restored by ex vivo treatment with the well-known HDACi SAHA.
Materials and methods
Chemical compounds
SAHA (SAHA, Merck, Kenilworth, NJ, USA) and Entinostat (MS275, Alexis Biochemicals, Roma, Italy) were dissolved in DMSO (Sigma-Aldrich, Milano, Italy) and used at 5 μm. Etoposide (Teva, Castleford, UK) was used at 34 μm; all-trans retinoic acid was used at 1 μm; valproic acid (Sigma-Aldrich) was used at 1 mm.
Cell line studies
U937, NB4, K562 and Molm-14 cells (DSMZ) were grown in RPMI 1640 medium (EuroClone) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), 1% glutamine (EuroClone), 1% penicillin/streptomycin (EuroClone) and 0.1% gentamycin (EuroClone), at 37 °C in air containing 5% CO2. KASUMI-1 (DSMZ) cells were grown similarly but with 20% fetal bovine serum (Sigma-Aldrich). HeLa and Kelly (DSMZ) cells were grown in DMEM medium (EuroClone) supplemented with the same components described above and in the same incubation conditions.
AML ex vivo samples
AML blasts and CD34+ cells were recovered from peripheral blood or bone marrow and purified by Ficoll density gradient separation (Sigma-Aldrich): after centrifugation at 1250 rpm for 25 min the layer of mononuclear cells was diluted in cell culture medium and further centrifuged for 5 min at the same speed. Cell pellet was dissolved in RPMI 1640 (EuroClone) enriched with 20% heat-inactivated fetal bovine serum (Sigma-Aldrich), 1% glutamine (EuroClone), 1% penicillin/streptomycin (EuroClone) and 0.1% gentamycin (EuroClone), and kept at 37 °C in air containing 5% CO2. AML blasts were treated with SAHA at 5 μm concentration for different times. All experiments were approved by the Second University of Naples ethical committee.
Purification and culture of mouse hematopoietic stem/progenitor cells
For in vitro experiments, total bone marrow was flushed out from femurs and tibia of 8-week-old wild-type C57BL/6 mice. The lineage-negative fraction was purified using the Lineage Cell Depletion Kit (Miltenyi Biotec, Calderara di Reno, Italy), according to manufacturer’s protocol. Lineage-depleted (Lin−) bone marrow progenitor cells were plated and cultured as previously described.16
DNase-seq analysis
DNase I libraries were prepared for UmiR-194-5p and Usc cells as described.17 In brief, nuclei were isolated and treated for 3 min with DNase I. The reaction was stopped with stop buffer (50 mm Tris-HCl pH 8, 100 mm NaCl, 0.10% SDS, 100 mm EDTA pH 8.0, 1 mm spermidine, 0.3 mm spermine). The sample was further fractionated on a 9% sucrose gradient at 25 000 rpm for 24 h at 16 °C. Fractions containing fragments smaller than 1 kb were purified and processed according to the Illumina library preparation protocol. Hotspots (DNAse I accessibility regions) were called as reported.18 Cutoff was set based on region size selection (>300 bp), ratio (three-fold) and tag total >30.
Additional details on cell culture, and molecular and cytological techniques used are provided in the Supplementary Methods.
Results
miR-194-5p expression is increased by the HDACi SAHA in AML cells
HDACi are currently used as chemosensitizers against cancer. To investigate the mode of action of HDAC sensitization in therapeutic treatment of leukemia cells, we assessed whether HDACi modulate miRNA expression in AMLs. We treated four AML cell lines (NB4, U937, K562 and KASUMI-1) with SAHA (5 μm, 6 h) and determined miRNA expression changes. We identified a cluster of 18 miRNAs commonly modified at least in two of the four cell lines (Supplementary Table S1). Two miRNAs, miR-194-5p and miR-92a-1* were identified as most significantly altered upon SAHA treatment and in all cell lines (P-value ⩽0.01); their expression was up- and downregulated, respectively, (Figure 1a; Supplementary Figure S1A). The expression level of several miRNAs was validated by quantitative PCR (data not shown). We focused on miR-194-5p as it showed a significant increase in response to SAHA in all four AML cell lines.
miR-194-5p is regulated by SAHA treatment in AML cell lines and regulates its new target, BCLAF1. (a) Fold change of microarray expression levels in log2 scale of two miRNAs commonly altered in four AML cell lines treated with SAHA compared to untreated control (P-value⩽0.01). (b) Venn diagram showing 119 downregulated genes after SAHA treatment in each AML cell line and 118 predicted target genes of miR-194-5p. BCLAF1 gene is the only downregulated predicted target gene. (c) Luciferase activity levels in U937 cells transfected with pGL3- BCLAF1 3'-UTR wild type (WT) and mutated (MUT) (1 μg), pre-miR-194-5p (100 nm) and pre-miR-scramble (100 nm) for 24 and 48 h. **P ⩽0.01; ***P ⩽0.001.
BCLAF1 is a new target of miR-194-5p
To identify targets of miR-194-5p, we interrogated three prediction databases (TargetScan, PicTar and miRanda), yielding a list of 119 commonly predicted potential target genes (Supplementary Table S2). We then performed gene expression microarray profiling and identified 240 genes commonly regulated in U937, NB4 and K562 cells upon SAHA treatment, 120 of which were upregulated and 118 downregulated. Intersecting the experimentally defined 118 commonly downregulated genes with the 119 predicted miR-194-5p target genes identified a single hit: BCLAF1 (Figure 1b). miR-194-5p and BCLAF1 expression in response to SAHA treatment was inversely correlated, as validated by quantitative PCR (Supplementary Figure S1B).
To determine whether BCLAF1 is a target of miR-194-5p in AMLs, we transiently overexpressed miR-194-5p in both U937 and NB4 cell lines. At 48 h after transfection, expression of BCLAF1 was strongly reduced upon miR-194-5p overexpression compared to miR-negative and carrier controls (Supplementary Figure S1C and D). We then cloned the predicted homologous seed region of miR-194-5p in the BCLAF1 3'-UTR in pGL3 control vector downstream of the luciferase gene and transfected the construct in U937 and HeLa cell lines. In these two different cell systems, a 60 and 90% reduction in luciferase activity was achieved, respectively (Figure 1c; Supplementary Figure S1E). The inverse correlation between miR-194-5p and BCLAF1 was further corroborated in SAHA-treated AML cells both at RNA and protein level at longer time (Supplementary Figures S1F and G). These results provide unambiguous evidence that miRNA-194-5p binds to the BCLAF1 3'-UTR to negatively regulate its expression.
miR-194-5p/BCLAF1 regulates cell cycle progression and apoptosis in AML cells
To elucidate the functional role of miR-194-5p, we mimicked SAHA-induced miR-194-5p expression through its stable re-expression in U937 cells (UmiR-194-5p). UmiR-194-5p-expressing lines (n=10) displayed negative regulation of BCLAF1 at both gene and protein level (Supplementary Figure S1H). All tested clones shared these features. Comparative genomic hybridization (CGH) array of UmiR-194-5p and scramble (Usc) cells did not show chromosomal rearrangements (Supplementary Figure S2A). UmiR-194-5p cells exhibited an altered global transcriptome compared to Usc cells (Supplementary Figures S2B and C; Supplementary Table S3).
Interestingly, UmiR-194-5p cells treated with SAHA displayed a large increase in the percentage of cells in pre-G1 phase (Figure 2a). The transcriptome upon SAHA treatment (Supplementary Figure S2D) highlighted regulation of apoptosis, RNA processing and biogenesis, regulation of myeloid cell differentiation and ribosome biogenesis (Supplementary Figure S2E; Supplementary Table S4). In agreement, Caspase 8 and 9 protein levels increased after SAHA stimulation (Supplementary Figure S2F), with a very concordant increase in Caspase 8 activity and an additional Caspase 9 activation (1.5- and 4.2-fold, respectively) (Figure 2b). These data were extended by the use of inhibitors of Caspase 9 (Z-LEHD-FMK) and 8 (Z-IETD-FMK). Blocking Caspase 9 activity in SAHA-treated cells completely abolished apoptosis in UmiR-194-5p cells, supporting a preponderant activation of Caspase 9-dependent pathways (Figure 2c). Increased reactive oxygen species production in SAHA-treated UmiR-194-5p cells corroborated the preferential activation of Caspase 9-dependent apoptosis (Supplementary Figure S2G). In agreement, re-expression of miR-194-5p in additional AML cells such as the Molm-14 cell line influences SAHA sensitivity inducing an increase of percentage of cells in pre-G1 phases (Supplementary Figure S2H). UmiR-194-5p cells also exhibited a better response to ‘classical’ chemotherapeutic agents such as etoposide (Supplementary Figure S2I). Taken together, our findings suggest a role for miR-194-5p in modulating drug response and susceptibility.
miR-194-5p/BCLAF1 module regulates cell cycle progression and apoptosis. (a) Pre-G1 cytofluorimetric analysis in UmiR-194-5p and Usc cells before and after SAHA treatment at 5 μm concentration for 24 h. (b) Cytofluorimetric analysis of Caspase 9 and 8 activity in UmiR-194-5p and Usc cells upon 24-h 5 μm SAHA treatment. Histogram showing percentage of Caspase 9 and 8 activity. (c) Pre-G1 cytofluorimetric analysis in UmiR-194-5p and Usc cells before and after 5 μm SAHA treatment with/without Z-LEHD-FMK and Z-IETD-FMK at 100 μm concentration for 40 h. FACS data are the result of three independent experiments each with three replicates, and are represented as ±s.e.m. *P ⩽0.05; **P ⩽0.01.
miR-194-5p/BCLAF1 controls dendritic differentiation commitment
Gene ontology (GO) analysis also indicated a role for miR-194-5p in differentiation, suggesting a potential link between AML differentiation block and miR-194-5p (Supplementary Figures S2B and C). To investigate this hypothesis, we treated Usc cells with different drugs known to preferentially induce differentiation: SAHA, valproic acid, all-trans retinoic acid and MS275 (also known as SNDX-275 or entinostat). We selected MS275 to evaluate the impact of miR-194-5p re-expression on AML cell differentiation as (i) it displayed higher differentiating potential (Supplementary Figure S3A) and (ii) is an HDACi reported for pro-differentiation action, as in some settings induce maturation before apoptosis.19, 20, 21
First, we verified morphological changes in UmiR-194-5p compared to Usc cells by measuring cell size (269.29 and 467.08 μm, respectively). Surprisingly, MS275 treatment of UmiR-194-5p cells resulted in a large increase in differentiated cells compared to Usc cells, as demonstrated by nitroblue tetrazolium assay (Supplementary Figure S3B). In agreement, UmiR-194-5p cells after MS275 treatment revealed features strongly reminiscent of dendritic cell morphology (Supplementary Figure S3C). Immunofluorescence microscopy of cytoskeletal actin filaments indicated that UmiR-194-5p cells form pluri-filaments of actin already after 5 min, actively polarizing F-actin at 15 min, and develop defined lamellipodia and filopodia structures at 6 and 24 h of MS275 treatment (Figure 3a).
miR-194-5p leads to dendritic differentiation commitment. (a) Inverted black and white images of IF of cytoskeletal actin filaments with Texas Red-labeled phalloidin antibody in UmiR-194-5p and Usc cells upon 5 μm MS275 treatment at 5 min, 15 min, 6 h and 24 h. Black bar scale: 10 μm. (b) Percentage of CD68-positive cells by manual cellular counting at confocal analysis of CD68 expression in UmiR-194-5p and Usc cells upon 24-h 5 μm MS275 treatment; cytofluorimetric assay for expression analysis of phycoerythrin-labeled CD80 and IL-12p70 secretion in UmiR-194-5p and Usc cells after MS275 treatment and in dendritic differentiation. (c) GO term classification of altered transcripts in UmiR-194-5p cells upon 24-h 5 μm MS275 treatment (FDR⩽0.05 and FC⩾2). FACS data are the result of three independent experiments each with three replicates, and are represented as ±s.e.m. **P ⩽0.01; ***P ⩽0.001. IL, interleukin.
The dendritic cell phenotype of UmiR-194-5p cells prompted us to investigate the expression of a human macrophage and dendritic marker, CD68. The number of CD68-expressing cells was higher in UmiR-194-5p than in Usc cells (47% and 10%, respectively), with 100% positive cells after MS275 treatment (Figure 3b). The specific dendritic marker CD80 was also expressed in 39% of UmiR-194-5p cells following MS275 treatment, whereas only 1.68% of Usc cells became CD80-positive (Figure 3b). Using granulocyte macrophage colony stimulating factor and interleukin-4, and stimulation with lipopolysaccharides medium to induce dendritic maturation,22 UmiR-194-5p but not Usc cells expressed CD80 and concomitantly secreted interleukin-12 (Figure 3b), thus strengthening the notion that miR-194-5p re-expression directs differentiation commitment to dendritic cells.
Analysis of transcriptome profiles revealed 80 activated and two downregulated genes upon MS275 treatment of UmiR-194-5p cells (Supplementary Table S5). GO analysis showed activation of genes involved in immune response via MHC class II, a distinct characteristic of dendritic differentiation (Figure 3c; Supplementary Table S6). Mechanistically, miR-194-5p-expressing cells downregulated BCLAF1, triggering an increase in reactive oxygen species (Supplementary Figure S2G) and higher Ca2+ production (Supplementary Figure S3D). Upon MS275 treatment, Ca2+ production was further increased. This state triggers activation of MHC class II gene promoters (Supplementary Figure S3E) and is also associated with higher expression and modulation of CIITA gene23 (Supplementary Figure S3F). Similarly to UmiR-194-5p cells, U937 BCLAF1 knockdown cells (UKDBCLAF1) do not display a significant alteration of cell cycle (Supplementary Figures S4A–C) but are more sensitive to SAHA treatment (Supplementary Figure S4D), showing a general reversion of the leukemic phenotype with drastically reduced growth and number of AML–colony-forming unit (Supplementary Figure S4E). In support, UKDBCLAF1 acquire features congruent to dendritic cell morphology, directly reminiscent of the UmiR-194-5p cells, even more after MS275 differentiation (Supplementary Figure S4F). In the opposite direction, BCLAF1 overexpression in U937 cells (UOverBCLAF1) increases proliferation and seem to lead to a certain resistance to SAHA (Supplementary Figures S4C and D). Finally, BCLAF1 actively enhanced tumorigenicity, as UOverBCLAF1 cells AML–colony-forming unit were increased in number and dimensions (Supplementary Figure S4E). Rescuing BCLAF1 expression in UmiR-194-5p reversed the effects induced by miR-194-5p overexpression, thus causally linking the connection of the miR-194-5p/BCLAF1 module (Supplementary Figures S5A–C). These features were commonly modulated also by different HDACi and other differentiating drugs (Supplementary Figure S5D).
Interestingly, by inhibiting miR-194-5p function using engineered U937-miArrest-194-5p (UmiR-194-5p-Arrest) cells, we corroborated miR-194-5p direct regulation of BCLAF1 and the effects on cell cycle progression (Supplementary Figures S6A and B). The majority of genes modulated by miR-194-5p re-expression displayed opposite trends in UmiR-194-5p-Arrest cells, suggesting a very specific role (and dosage effect) for miR-194-5p in controlling differentiation (Supplementary Figure S6C). Specific miR-194-5p repression in AMLs appears to be closely linked to maturation block, a characteristic of acute leukemias. Taken together, these data show that miR-194-5p via targeting BCLAF1 commits cells to dendritic cell differentiation.
miR-194-5p/BCLAF1 impacts on chromatin accessibility in AML
Evidence that miR-194-5p/BCLAF1 equilibrium impacts on maturation, cell fate and susceptibility to anticancer treatment, together with the suggested role of BCLAF1 as a transcription factor,24 led us to speculate that the miR-194-5p/BCLAF1 module might affect chromatin state. Although the karyotype of UmiR-194-5p cells did not reveal significant chromosomal rearrangements (Supplementary Figure S2A), a substantial increase in K9/K14 and K27 acetylation on histone 3 was detected (Figure 4a). This increase corresponded to a major shift in chromatin accessibility between UmiR-194-5p and Usc cells, with 14.126 accessible regions in Usc cells (Usc accessible) showing reduced accessibility and 8.169 closed regions becoming accessible in U937 cells expressing miR-194-5p (UmiR-194-5p accessible) (Figure 4b; Supplementary Figure S7C). Genomic annotation of chromatin accessible sites in UmiR-194-5p and Usc cells revealed 25% of sites located in promoter regions (Figure 4c).
miR-194-5p establishes a new euchromatin arrangement. (a) Western blot showing expression levels of H3K9/K14ac analysed in UmiR-194-5p and Usc cells upon 24-h 5 μm SAHA or MS275 treatment. Normalization with H4. (b) Differential DNase I accessible regions in UmiR-194-5p and Usc cells. The heat map shows tag counts in 500 bp bins around accessible sites specifically identified in Usc cells (top) or in UmiR-194-5p cells (bottom). (c) Genomic localization of DNase I accessible regions in UmiR-194-5p and Usc cells, represented as % of peaks.
Overlap** the DNase I accessible regions to BCLAF1-binding sites (GEO:GSM803515;GEO:GSM803509, ENCODE), a reduction of BCLAF1 accessible binding regions was detected in the UmiR-194-5p cells with respect to Usc, 12% vs 31%, respectively (Supplementary Figure S7D). BCLAF1-associated regions preferentially localized at promoters in UmiR-194-5p, independent on the expression directionality (Supplementary Figures S7E and F). These results corroborate and strengthen the miR-194-5p/BCLAF1 module impact on chromatin accessibility.
BCLAF1 functions via an HDAC4-dependent pathway in AML
To clarify the implication of BCLAF1 regulation by miR-194-5p in tumorigenesis and therapeutic response to HDACi in AMLs, we investigated the cellular localization of BCLAF1. In AML cells, BCLAF1 was localized in dot-like structures throughout the nucleus25, 26 (Figure 5a). In contrast, upon miR-194-5p (re)expression, residual BCLAF1 shuttled from nucleus to cytosol (Figure 5a). Such shuttling was also observed upon the maturation burst induced by MS275, with BCLAF1 present in both cytosol and nucleus. Conversely, SAHA treatment (associated with a preferential apoptotic response) retained BCLAF1 in nucleus (Figure 5a).
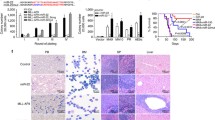
Intracellular localization of BCLAF1 determines its differential function. (a) IF analysis of BCLAF1 expression and localization in Usc and UmiR-194-5p cells upon 5 μm MS275 or SAHA treatment at 6 and 24 h. Black bar scale: 10 μm. (b) BCLAF1 and miR-194-5p expression by RNA-seq reads in human hematopoietic progenitors. (c) Expression levels of miR-194-5p and BCLAF1 analysed by real-time PCR during ex vivo expansion of lineage-depleted mouse bone morrow cells at T0, T1, T2 and T3 (0, 1, 2 and 4 days from bone marrow isolation); IF analysis of BCLAF1 localization during lineage differentiation of normal bone marrow mouse progenitors at T1, T2, T3 (1, 2, 4 days from bone marrow isolation) and again at T2 upon 24-h 5 μm MS275 or SAHA treatment. *P ⩽0.05.
In support of these finding, we verified at transcriptional level during lineage commitment of human blood progenitors27 that miR-194-5p expression was low and correlated with increased Bclaf1 expression in hematopoietic progenitors (Figure 5b). Besides, upon cell culture of freshly isolated murine Lin− murine bone marrow progenitor cells (T0) into CD34+-enriched hematopoietic cells (T1) and more differentiated stages (T2, T3), miR-194-5p expression increased and Bclaf1 levels decreased (Figure 5c). Ex vivo culturing of Lin− murine bone marrow progenitor cells (LIN-CD150+) reduces their differentiative potential, inducing a more differentiated state (LIN-CD150−) (Supplementary References 4 and 5). In a time course analysis from T0 to T3, the correlation between miR-194-5p/BCLAF1 module expression and reduction multi-lineage differentiation potential of Lin− murine bone marrow progenitor cells was demonstrated (Figure 5c; Supplementary Figure S8A). Noteworthy, in normal mouse CD34+-enriched myeloid progenitors, BCLAF1 is located at peri-membrane and not in nuclei, whereas in AMLs it is nuclear, suggesting that its expression and location are functionally altered. In normal progenitors, the MS275-induced differentiation burst maintained peri-membrane cytosol location, whereas SAHA treatment led to nuclear location (Figure 5c). Accordingly, we found BCLAF1 location in human CD34+ progenitor cells in the cytosol and human primary AML blasts in the nucleus (Supplementary Figure S8B).
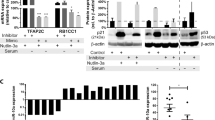
Thus, we hypothesized that the differential BCLAF1 localization observed might be causally linked to the differential HDAC targeting exerted by SAHA or MS275. Several class II HDACs are reported to shuttle between nucleus and cytoplasm,28, 29 and our observation of HDAC4 expression in UmiR-194-5p cells strengthens this hypothesis. Excitingly, unlike all other HDACs, HDAC4 was selectively transcriptionally upregulated (Figure 6a), also for direct regulation of accessibility on its promoter region (Figure 6b). miR-194-5p seems to regulate specific members of MAPK pathway, such as CAMKII and 14-3-3 at chromatin (data not shown) and protein level (Supplementary Figure S7A). Upon treatment with the two different HDACi, HDAC4 consistently co-localized with BCLAF1 (Supplementary Figure S7B): in UmiR-194-5p cells, HDAC4 (and its phosphorylated form) was mainly located in cytoplasm, whereas upon SAHA treatment HDAC4 partially shuttled into nucleus.
BCLAF1 functions via an HDAC4-dependent pathway in AML. (a) Heat map of gene microarray expression in log2 scale of HDAC family members in UmiR-194-5p and Usc cells. (b) DNase I profile of HDAC4 accessible region in UmiR-194-5p and Usc cells. (c) Protein immunoprecipitation of BCLAF1 and HDAC4 in Usc and UmiR-194-5p cells. Western blot shows binding for CaMKII, CaMKII-P and HDAC4-P. IF analysis of BCLAF1 localization in HDAC4-silenced cell (UshHDAC4) vs appropriate control (Uscsh), before and after transfection with miR-194-5p to stimulate the BCLAF1 shuttling from nucleus into the cytosol. This phenomena was not observed in UshHDAC4.
In UmiR-194-5p cells, HDAC4 and HDAC4-P, but also CaMKII, CaMKII-P and 14-3-3-P, were mainly found in cytosol (Supplementary Figure S7B). Examination of the sequences surrounding the conserved phosphorylation residues in the class IIa HDACs revealed that they are closely related to the consensus phosphorylation sites of CaMK.23 We hypothesized that HDAC4 (and its phosphorylated form) might bind BCLAF1, possibly in a complex with CaMKII. In support, immunoprecipitation of BCLAF1 showed binding with HDAC4-P and CaMKII-P. Similarly, HDAC4 was able to form endogenous complexes with BCLAF1 and CaMKII (Figure 6c). BCLAF1 differential location was HDAC4-dependent. Indeed, HDAC4 silencing (UshHDAC4) caused the absence of BCLAF1 cytosolic shuttling in UshHDAC4miR-194-5p cells, with respect to control (Uscsh) (Figure 6c). The fact that expression of both HDAC4 and CaMKII can be reduced by applying miArrest technology strengthens the importance of the modulatory role of miR-194-5p (Supplementary Figure S6D). In U937-miArrest-194-5p cells, inhibition of miR-194-5p function showed an opposite trend in all the newly identified targets (Supplementary Figures S6C–E). These findings suggest that an unbalanced dose of miR-194-5p triggers the kinase pathway involving BCLAF1, HDAC4 and CaMKII, modulating either differentiation and/or apoptosis.
miR-194-5p/BCLAF1 deregulation is a hallmark of AML tumorigenesis
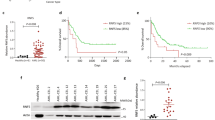
The emerging role for BCLAF1 (and possibly miR-194-5p) in AML pathogenesis and/or progression is further supported by the high expression of BCLAF1 in chronic lymphocytic leukemia, AML and neuroblastoma (Supplementary Figures S9A–C). miR-194-5p/BCLAF1 inversely correlated expression was assessed ex vivo in normal human CD34+ myeloid progenitors and AML samples. Importantly, normal progenitors invariably displayed higher levels of miR-194-5p compared to primary AML blasts independently of karyotype and immunophenotype (n=60) (Supplementary Table S7), suggesting a general repression of miR-194-5p and exertion of its function during leukemogenesis (Figure 7a; Supplementary Figure S9A; Supplementary Table S8). BCLAF1 showed the opposite pattern, displaying higher expression in AML and lower in CD34+ cells (Figure 7a; Supplementary Figure S9D; Supplementary Table S8). Treatment of AML blasts ex vivo with SAHA reverted the expression of miR-194-5p/BCLAF1 to a level more comparable to normal cells (Figure 7a; Supplementary Figure S9D; Supplementary Table S8). Comparative analysis of miR-194-5p/BCLAF1 in CD34+ and AML cells confirmed a highly significant direct inverse correlation between expression and cancer phenotype (Figure 7a; Supplementary Figure S9D). High BCLAF1 expression in AML was further confirmed in an analysis of 200 clinically annotated adult cases of de novo AML30 (Supplementary Figure S9E). We also investigated the induction of cell death in 41 samples from AML patient’s blasts. Thirty-seven out of 41 blast samples were responsive to SAHA treatment with an apoptosis fold induction higher than two, whereas five samples were not responsive (Supplementary Table S9). Among all responsive AML samples, 56.76% displayed inverse correlation between miR-194-5p and BCLAF1 expression levels, and their modulation upon SAHA treatment (Figure 7b). By merging data with the medical outcome information (available only on 44 out of 61 samples), higher miR-194-5p levels (and the lower expression of BCLAF1) correlated to favorable prognosis based on the 5-year survival rate (Figure 7c). No predominant correlation subgroups were detected in the not-responsive category (Supplementary Figure S9F), although miR-194-5p expression level was significantly lower if compared to the responsive group (Supplementary Figure S9G; Supplementary Table 10). Finally, miR-194-5p and BCLAF1 module in AML and its regulation by SAHA was corroborated at gene and protein level in vivo in human AML xenograft mouse model (Supplementary Figure S9H).
Inverse correlation between miR-194-5p/BCLAF1 is maintained in ex vivo AML blasts. (a) Expression levels of miR-194-5p and BCLAF1 analysed by RT-PCR in AML patients before and after 5 μm SAHA treatment at different times. Student’s t-test analysis performed between each patient group before and after SAHA treatment. (b) Percentages of AML patients responsive (R) and not responsive (NR) to SAHA treatment based on a threshold of two for apoptosis fold induction. Double pie chart showing subpopulations of responsive patients based on expression levels of miR-194-5p and BCLAF1. RT-PCR data are the result of three independent experiments each with three replicates, and are represented as ±s.e.m. RNU6B and GAPDH were used for data normalization for miRNA and gene expression. (c) Relative expression levels of miR-194-5p and BCLAF1 analysed by RT-PCR in untreated AML patient’s vs CD34+ samples. The histogram reports the media of miR-194-5p/BCLAF1 expression correlated to the 5-year survival, dividing the patients in two categories, as indicated. t-test analysis was performed and P-values are reported in the graph. RT, real-time.
Altogether, these data hint at a role for miR-194-5p/BCLAF1 in defining tumorigenic arrangement and mediating sensitivity to SAHA treatment.
Discussion
Epigenetic mechanisms regulate the miRNA expression and, conversely, subsets of miRNAs control important epigenetic regulators, establishing a regulatory circuit to stabilize and modulate gene expression patterns.31, 32 The link between miRNA deregulation and malignancies has been clearly defined.3 Some reports identified ‘epidrug’-responsive miRNAs whose expression was affected by ‘epi’-treatment in different cancers.33, 34 However, insights into miRNA function in cell behavior, differentiation, stemness and pathology, and their mode of action have so far been scarce.
We provide evidence that miR-194-5p exhibits an oncosuppressor role and a key function in cell reactivity, drug response and differentiation commitment. A body of literature suggests that the role of miR-194-5p (mainly its aberrant expression) depends on the context and/or dynamics of different targets within a specific cell.35, 36, 37, 38
We investigated whether miR-194-5p and its respective target(s) might regulate the equilibrium of biological processes such as maturation/differentiation and pathogenesis. We found that miR-194-5p acts by negatively regulating BCLAF1, a newly identified target gene. In human AML cell lines and in a statistically significant number of human primary AML blasts compared to normal myeloid progenitors, expression of miR-194-5p (low) and BCLAF1 (high) is inversely correlated, defining leukemic phenotype. This finding suggests compelling evidence that miR-194-5p might play a tumor suppressor role, and that its expression level in AMLs may have preclinical significance. We also found that SAHA upregulates miR-194-5p in all AML models tested including ex vivo treatment of human primary blasts and that miR-194-5p re-expression leads to chemo-or epi-drugs sensitization, restoring the physiological-like state. Chip-seq data suggest an active epigenetic chromatin regulation by SAHA treatment on putative miR-194-5p promoter (Supplementary Figures S8D and E). Specifically, miR-194-5p re-expression sensitizes cells to SAHA-induced pro-apoptotic effects through a Caspase 9-dependent pathway and by functional determination of BCLAF1 nuclear localization. Conversely, MS275 treatment affects the ability of miR-194-5p to induce differentiation, most likely by affecting cytosolic localization of BCLAF1.
Very recently, miR-194-5p was reported as a candidate biomarker for myelodysplastic syndromes, being its low expression associated with poor survival in myelodysplastic syndromes.39 To date, few reports have described the involvement of miR-194-5p40, 41 and BCLAF125, 26 in differentiation. As acute leukemias represent the prototype of a cancer caused by so-called ‘cancer stem cells’, where all blasts have a marked deficit in maturation, we hypothesize that miR-194-5p acts as a checkpoint for stem cells to decide on their fate. Supporting and strengthening our suppositions, the miR-194-5p/BCLAF1 module is similarly deregulated in neuroblastoma, a cancer in which cells have impaired maturation. miR-194-5p may thus perform the role of ‘commitment maker’.
Interestingly, our findings underscore that re-establishment of miR-194-5p expression levels also permits differentiation toward fully specialized cells, in our case to dendritic cells. The effect of miR-194-5p on potential maturation into dendritic cells suggests a role in immune response in AML by myeloid differentiation commitment.
It remains to be clarified whether miR-194-5p deregulation is an initial event or is part of leukemogenesis progression. Recent studies have highlighted the role of miRNAs in enhancing sensitivity to traditional therapies.13, 42 Here, we show a mechanism of action exerted by miR-194-5p in association with BCLAF1.
BCLAF1 was originally identified as a death-inducing repressor,24 but subsequent reports indicated a much wider function, not necessarily linked to BCL2.26 For example, BCLAF1 is a γH2AX-interacting tumor suppressor, Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233. Gangaraju VK, Lin H . MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol 2009; 10: 116–125. Lieberman J, Slack F, Pandolfi PP, Chinnaiyan A, Agami R, Mendell JT . Noncoding RNAs and cancer. Cell 2013; 153: 9–10. Song SJ, Pandolfi PP . MicroRNAs in the pathogenesis of myelodysplastic syndromes and myeloid leukaemia. Curr Opin Hematol 2014; 21: 276–282. Dell’aversana C, Altucci L . miRNA-mediated deregulation in leukemia. Front Genet 2012; 3: 252. Estey E, Dohner H . Acute myeloid leukaemia. Lancet 2006; 368: 1894–1907. Lowenberg B, Downing JR, Burnett A . Acute myeloid leukemia. N Engl J Med 1999; 341: 1051–1062. Bolden JE, Peart MJ, Johnstone RW . Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 2006; 5: 769–784. Frew AJ, Johnstone RW, Bolden JE . Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett 2009; 280: 125–133. Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 2009; 125: 2737–2743. Shin S, Lee EM, Cha HJ, Bae S, Jung JH, Lee SM et al. MicroRNAs that respond to histone deacetylase inhibitor SAHA and p53 in HCT116 human colon carcinoma cells. Int J Oncol 2009; 35: 1343–1352. Saito Y, Suzuki H, Tsugawa H, Nakagawa I, Matsuzaki J, Kanai Y et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene 2009; 28: 2738–2744. Lepore I, Dell'Aversana C, Pilyugin M, Conte M, Nebbioso A, De Bellis F et al. HDAC inhibitors repress BARD1 isoform expression in acute myeloid leukemia cells via activation of miR-19a and/or b. PLoS One 2013; 8: e83018. Ageberg M, Rydstrom K, Relander T, Drott K . The histone deacetylase inhibitor valproic acid sensitizes diffuse large B-cell lymphoma cell lines to CHOP-induced cell death. Am J Transl Res 2013; 5: 170–183. Perego P, Zuco V, Gatti L, Zunino F . Sensitization of tumor cells by targeting histone deacetylases. Biochem Pharmacol 2012; 83: 987–994. Zito E, Buono M, Pepe S, Settembre C, Annunziata I, Surace EM et al. Sulfatase modifying factor 1 trafficking through the cells: from endoplasmic reticulum to the endoplasmic reticulum. EMBO J 2007; 26: 2443–2453. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014; 345: 1251086. Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA . Circuitry and dynamics of human transcription factor regulatory networks. Cell 2012; 150: 1274–1286. D’Amato L, Dell’Aversana C, Conte M, Ciotta A, Scisciola L, Carissimo A et al. ARHGEF3 controls HDACi-induced differentiation via RhoA-dependent pathways in acute myeloid leukemias. Epigenetics 2015; 10: 6–18. Rosato RR, Almenara JA, Grant S . The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer Res 2003; 63: 3637–3645. Vrana JA, Decker RH, Johnson CR, Wang Z, Jarvis WD, Richon VM et al. Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene 1999; 18: 7016–7025. Bontkes HJ, De Gruijl TD, Schuurhuis GJ, Scheper RJ, Meijer CJ, Hooijberg E . Expansion of dendritic cell precursors from human CD34(+) progenitor cells isolated from healthy donor blood; growth factor combination determines proliferation rate and functional outcome. J Leukoc Biol 2002; 72: 321–329. McKinsey TA, Kuwahara K, Bezprozvannaya S, Olson EN . Class II histone deacetylases confer signal responsiveness to the ankyrin-repeat proteins ANKRA2 and RFXANK. Mol Biol Cell 2006; 17: 438–447. Kasof GM, Goyal L, White E . Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol 1999; 19: 4390–4404. McPherson JP, Sarras H, Lemmers B, Tamblyn L, Migon E, Matysiak-Zablocki E et al. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ 2009; 16: 331–339. Orieux G, Picault L, Slembrouck A, Roger JE, Guillonneau X, Sahel JA et al. Involvement of Bcl-2-associated transcription factor 1 in the differentiation of early-born retinal cells. J Neurosci 2014; 34: 1530–1541. Chen L, Kostadima M, Martens JH, Canu G, Garcia SP, Turro E et al. Transcriptional diversity during lineage commitment of human blood progenitors. Science 2014; 345: 1251033. Verdin E, Dequiedt F, Kasler HG . Class II histone deacetylases: versatile regulators. Trends Genet 2003; 19: 286–293. Delcuve GP, Khan DH, Davie JR . Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenet 2012; 4: 5. Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. Dai E, Yu X, Zhang Y, Meng F, Wang S, Liu X et al. EpimiR: a database of curated mutual regulation between miRNAs and epigenetic modifications. Database (Oxford) 2014; 2014: bau023. Sato F, Tsuchiya S, Meltzer SJ, Shimizu K . MicroRNAs and epigenetics. FEBS J 2011; 278: 1598–1609. Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S . Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE 2011; 6: e24099. Lee EM, Shin S, Cha HJ, Yoon Y, Bae S, Jung JH et al. Suberoylanilide hydroxamic acid (SAHA) changes microRNA expression profiles in A549 human non-small cell lung cancer cells. Int J Mol Med 2009; 24: 45–50. Han K, Zhao T, Chen X, Bian N, Yang T, Ma Q et al. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol 2014; 45: 1437–1449. Harris-Arnold A, Arnold CP, Schaffert S, Hatton O, Krams SM, Esquivel CO et al. Epstein-Barr virus modulates host cell microRNA-194 to promote IL-10 production and B lymphoma cell survival. Am J Transplant 2015; 15: 2814–2824. Khella HW, Bakhet M, Allo G, Jewett MA, Girgis AH, Latif A et al. miR-192, miR-194 and miR-215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 2013; 34: 2231–2239. Wu X, Liu T, Fang O, Leach LJ, Hu X, Luo Z . miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27(kip1). Oncogene 2014; 33: 1506–1514. Choi JS, Nam MH, Yoon SY, Kang SH . MicroRNA-194-5p could serve as a diagnostic and prognostic biomarker in myelodysplastic syndromes. Leuk Res 2015; 39: 763–768. Xu J, Kang Y, Liao WM, Yu L . MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS ONE 2012; 7: e31861. Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA 2008; 14: 1433–1442. Sampath D, Malik A, Plunkett W, Nowak B, Williams B, Burton M et al. Phase I clinical, pharmacokinetic, and pharmacodynamic study of the Akt-inhibitor triciribine phosphate monohydrate in patients with advanced hematologic malignancies. Leuk Res 2013; 37: 1461–1467. Lee YY, Yu YB, Gunawardena HP, **e L, Chen X . BCLAF1 is a radiation-induced H2AX-interacting partner involved in gammaH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Dis 2012; 3: e359. Liu H, Lu ZG, Miki Y, Yoshida K . Protein kinase C delta induces transcription of the TP53 tumor suppressor gene by controlling death-promoting factor Btf in the apoptotic response to DNA damage. Mol Cell Biol 2007; 27: 8480–8491. Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell 2014; 54: 445–459. Nishino TG, Miyazaki M, Hoshino H, Miwa Y, Horinouchi S, Yoshida M . 14-3-3 regulates the nuclear import of class IIa histone deacetylases. Biochem Biophys Res Commun 2008; 377: 852–856. Awad S, Kunhi M, Little GH, Bai Y, An W, Bers D et al. Nuclear CaMKII enhances histone H3 phosphorylation and remodels chromatin during cardiac hypertrophy. Nucleic Acids Res 2013; 41: 7656–7672. Hohl M, Wagner M, Reil JC, Muller SA, Tauchnitz M, Zimmer AM et al. HDAC4 controls histone methylation in response to elevated cardiac load. J Clin Invest 2013; 123: 1359–1370. Liu F, Dowling M, Yang XJ, Kao GD . Caspase-mediated specific cleavage of human histone deacetylase 4. J Biol Chem 2004; 279: 34537–34546. Seeger RC . Immunology and immunotherapy of neuroblastoma. Semin Cancer Biol 2011; 21: 229–237. Anguille S, Van Tendeloo VF, Berneman ZN . Leukemia-associated antigens and their relevance to the immunotherapy of acute myeloid leukemia. Leukemia 2012; 26: 2186–2196. We thank S Arbucci (Microscopy, IGB) for technical assistance; T Giuliano (NGS, TIGEM) for CGH; M Conte, I Lepore for help during conducting experiments; C Fisher for linguistic editing. Grants: Blueprint (282510), EPIGEN (MIUR-CNR); MIUR (PRIN2012ZHN9YH; 20152TE5PK), AIRC (17217); POR Campania FESR 2007-2013-MOVIE (B25C13000240007), COST EPICHEMBIO CM1406. Accession numbers: Microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession numbers: GSE37374, GSE69959, GSE73656 and GSE55154. GEO accession number of DNAse I data is GSE76426.
Author contributions
Carmela Dell’Aversana: performed the majority of experiments and wrote the manuscript. Cristina Giorgio, Loredana D’Amato, Antonella Di Costanzo and Gabriella Lania: performed cell-based experiments. Filomena Matarese, Sadia Saeed: performed DNAse hypersensitivity assays. Saverio Minucci, Isabella Pallavicini: analysed the murine model. Valeria Belsito Petrizzi, Concetta Ingenito: analysed the clinical data. Annamaria Carissimo, Joost HA Martens, Hendrik G Stunnenberg: analysed ‘omics’ and data interpretation. Lucia Altucci: conceived the study and wrote the manuscript. The authors declare no conflict of interest. Supplementary Information accompanies this paper on the Leukemia website This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/ Dell'Aversana, C., Giorgio, C., D'Amato, L. et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis.
Leukemia 31, 2315–2325 (2017). https://doi.org/10.1038/leu.2017.64 Received: Revised: Accepted: Published: Issue Date: DOI: https://doi.org/10.1038/leu.2017.64 Biochemical Genetics (2024) Medical Oncology (2024) Cellular and Molecular Life Sciences (2024) Biochemical Genetics (2024) Cancer Immunology, Immunotherapy (2023)References
Acknowledgements
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Additional information
Rights and permissions
About this article
Cite this article
This article is cited by
Pan-Cancer Analysis Identifies BCLAF1 as a Potential Biomarker for Renal Cell Carcinoma
A novel therapeutic strategy: the significance of exosomal miRNAs in acute myeloid leukemia
BCLAF1 binds SPOP to stabilize PD-L1 and promotes the development and immune escape of hepatocellular carcinoma
BCLAF1 is Expressed as a Potential Anti-oncogene in Bile Duct Cancer
BCLAF1-induced HIF-1α accumulation under normoxia enhances PD-L1 treatment resistances via BCLAF1-CUL3 complex