Abstract
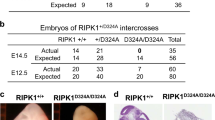
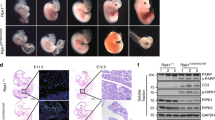
RIP1 is an essential regulator of TNF-induced signaling complexes mediating NF-κB activation, apoptosis and necroptosis. Loss of Rip1 rescues the embryonic lethality of Fadd or Caspase-8-deficient mice, even though the double knockout mice die shortly after birth like Rip1-deficient mice. Recent studies demonstrated that mice expressing RIP1 kinase-dead mutants developed normally and resisted necroptotic stimuli in vitro and in vivo. However, the impact of RIP1 kinase activity on Fadd−/− embryonic development remains unknown. Here, we engineered two RIP1 kinase inactive mutant mouse lines, a Rip1K45A/K45A mouse line as previously reported and a novel Rip1Δ/Δ mouse line with an altered P-loop in the kinase domain. While RIP1K45A could not rescue the embryonic lethality of Fadd-deficient mice at E11.5, RIP1Δ rescued lethality of Fadd−/− mice at E11.5 and Fadd−/−Rip1Δ/Δ mice eventually died at E16.5 due to excessive death of fetal liver cells and unregulated inflammation. Under necropotosis-inducing conditions, comparing to Rip1K45A/K45A cells, Rip1Δ/Δcells displayed reduced phosphorylation and oligomerization of RIP3 and MLKL, which lead to increased cell viability. Thus, our study provides genetic evidence that different kinase inactive mutations have distinct impacts on the embryogenesis of Fadd-deficient mice, which might attribute to their extents of protection on necroptosis signaling.
Similar content being viewed by others
Main
The receptor-interacting protein kinase 1 (RIP1) functions as a key regulator for NF-κB activation, apoptosis and necroptosis induced by tumor necrosis factor (TNF-α).1, TNF-α and z-VAD were purchased from R&D (Minneapolis, MN, USA) and Calbiochem (Anaheim, CA, USA), respectively. The Smac mimetic, Cycloheximide and LPS were obtained from Sigma. Nec-1 was from Enzo Life Science (Alexis, USA). The following antibodies were used for western blotting: p-IkBa (Cell Signaling), p-ERK (Cell Signaling Technology, Danvers, MA, USA), ERK (Cell Signaling), p-P38 (Cell Signaling), P38 (Cell Signaling), p-P65 (Cell Signaling), p-JNK (Cell Signaling), JNK (Cell Signaling), PARP (Cell Signaling), Caspase-3 (Cell Signaling), HA (Cell Signaling), Caspase-8 (Enzo Life Science), RIP1 (BD Biosciences, Franklin Lakes, NJ, USA), p-RIP1(S166) (Cell Signaling) mouse RIP3 (ProSci, San Diego, CA, USA), MLKL (Abcam, Cambridge, UK), p-MLKL (Abcam), β-actin, α-Tubulin and anti-flag-HRP (Sigma). Anti-phospho-RIP3 antibody (mouse) was generated in our lab and immunoaffinity-purified. Cell viability was determined by measuring ATP levels using Cell Titer-Glo kit (Promega, Madison, WI, USA). Mice were housed in a specific pathogen-free facility, which belongs to Institute for Nutritional Sciences. Fadd−/− mice (C57BL/6) were gifted by Dr. Jianke Zhang (Thomas Jefferson University, Philadelphia, USA), and Rip3−/− mice (C57BL/6) were provided by Dr. **aodong Wang (NIBS, Bei**g, China). Animals were subsequently backcrossed on a C57BL/6 background for at least 10 generations. A novel mutant, RIP1Δ, was obtained from the experiment with two amino acids G26F27 in the P-loop of RIP1 deleted. To generate Rip1Δ/Δ, Rip1K45A/K45A, Rip1−/− mice by crispr-cas9 mutation system (Bioray Laboratories Inc., Shanghai, China), different sgRNA were designed to target RIP1 kinase domain. (Rip1Δ/Δmutant with gRNA: 5′-GACCTAGACAGCGGAGGCTT-3′; Rip1K45A/K45A mutant with gRNA: 5′-GCCCTGTGTATACTTTTTTC-3′; Rip1−/− with gRNA: 5′-GATGGCATCCAGTGACCTGC-3′). Additional information is provided upon request. All mutant mice and WT mice used in these studies shared a common genetic C57BL/6 background. Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (CAS), University of Chinese Academy of Sciences. Male WT, Rip3−/− and Rip1Δ/Δ mice littermates at 8 weeks of age (n=7) were treated every hour for 12 consecutive hours with cerulein (50 μg/kg, Sigma) intraperitoneal injection. Animals were assayed 24 h after the first injection. Serum amylase activity was assayed by amylase activity assay kit (Sigma). MDFs were separated from the skin of newborn mice, and MEFs were isolated from E13.5 to E14.5 embryos. MDFs and MEFs were cultured in DMEM medium supplemented with 10% FBS and penicillin/streptomycin. BMDMs from isolated bone marrow cells collected from mouse femurs and tibias were induced to differentiate in vitro. Bone marrow cells were cultured for 7 days in RPMI medium containing 10% FBS, penicillin/streptomycin and 50 ng/ml M-CSF, and medium was changed every 2 days. Thymocytes were isolated from thymus and cultured in RPMI medium containing 10% FBS, penicillin/streptomycin and 50 μM β-ME. Cells were harvested at different time points, washed with PBS and lysates with 1 × SDS sample buffer containing 100 mM DTT and boiled for 5 min at 95 °C for reducing gel. For mouse tissue protein extraction, E14.5 fetal liver and other tissues were ground by pestle and mortar with liquid N2, and the protein was extracted with RIPA lysis buffer. The lysates were cleared by centrifugation for 20 min at 13 200 × g, quantified by BCA kit (Thermo Scientific, Rockford, IL, USA) then mixed with SDS sample buffer and boiled at 95 °C for 5 min. The samples were separated using SDS-PAGE, transferred to PVDF membrane (Millipore, Darmstadt, Germany) with 100 v for 2 h. The proteins were detected by using a chemiluminescent substrate (Thermo Scientific). To immunoprecipitate RIP1, cell extract protein was incubated for 3 h with 5 μl of RIP1 antibody (BD Biosciences). After mixing end over end for overnight (4 °C) with 30 μl of G-Agarose beads, the agarose was collected and washed three times with cell lysis buffer (Tris-HCl 20 mmol/l (pH 7.5), NaCl 150 mM, EDTA 1 mM, EGTA 1 mM, Triton X-100 1%, Sodium pyrophosphate 2.5 mM, β-Glycerrophosphate 1 mM, NaVO4 1 mM, Leupeptin 1 μg/ml.). Immunoprecipitates were denatured in SDS, subjected to SDS-PAGE, and immunoblotted. The cells were cultured in six-well plates and treated with indicated stimuli. Cells were harvested at different time points and lysed with 2 × DTT-free sample buffer (Tris-Cl (PH 6.8) 125 mM, SDS 4%, Glycerol 20%, Bromophenol blue 0.02%) immediately. Total cell lysates were separated using SDS-PAGE, and transferred to PVDF membrane (Millipore), and western blotting was performed with RIP3 or MLKL antibodies. Antibodies against mouse CD3, CD4, CD8, CD19, Mac-1 and Gr-1 from eBioscience (Carlsbad, CA, USA) were fluorescence-conjugated and were used for flow cytometry analysis in this study. We prepared single-cell suspension from lymph nodes, spleen and thymus, respectively, and stained them with fluorescence-conjugated antibodies for half an hour in staining buffer. After staining, cells were immediately analyzed by flow cytometry (FACSAria III, BD Biosciences). MDFs were plated overnight on coverslips before various stimulations. After stimulation, cells were washed with PBS and fixed with 4% PFA in PBS for 15 min. Next, the cells were blocked with 0.3% Triton X-100 and 5% normal donkey serum (Jackson immunoResearch, Baltimore Pike, West Grove, PA, USA) for 1 h at room temperature, then followed by first antibody incubation at 4 °C for overnight. Signals were developed with Alexa fluorescence antibodies (Invitrogen). Finally, the cells were stained with DAPI for 10 min. Confocal microscopy analysis was performed using a Zeiss 710 laser-scanning microscope (Zeiss, Thornwood, NY, USA). Total RNA was extracted using Trizol reagent (Life Technologies), according to the manufacturer’s instructions. After quantification, 2 μg total RNA was reverse transcribed to complementary DNA (Takara, Dalian, China). Transcript levels of indicated cytokines were quantified by quantitative RT-PCR on an ABI 7500 real-time PCR instrument with SYBR Green. Relative expression was calculated using LC32 as an internal control as indicated. Primers used were as follows: mIL-1b: 5′-CCCAACTGGTACATCAGCAC-3′ and 5′-TCTGCTCATTCACGAAAAGG-3′; mTNF: 5′-CCCACTCTGACCCCTTTACT-3′ and 5′-TTTGAGTCCTTGATGGTGGT-3′; mIL-6: 5′-CGGAGAGGAGACTTCACAGA-3′ and 5′-CCAGTTTGGTAGCATCCATC-3′; mCXCL-1: 5′-CTGGGATTCACCTCAAGAACATC-3′ and 5′-CAGGGTCAAGGCAAGCCTC-3′; mMCP-1: 5′-TTAAAAACCTGGATCGGAACCAA-3′ and 5′-GCATTAGCTTCAGATTTACGGGT-3′; mCCL-5:5′-GCTGCTTTGCCTACCTCTCC-3′ and 5′-TCGAGTGACAAACACGACTGC-3′.Materials and Methods
Reagents
Mice
Cerulein-induced acute pancreatitis
Isolation and culture of MDFs, MEFs, BMDMs and thymocytes
Immunoblotting and immunoprecipitation
RIP3 and MLKL oligomerization detection
Flow cytometry
Immunofluorescence
RT-PCR
References
Stanger BZ, Leder P, Lee TH, Kim E, Seed B . RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell 1995; 81: 513–523.
Hsu H, **ong J, Goeddel DV . The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 1995; 81: 495–504.
Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P . RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell Death Differ 2007; 14: 400–410.
Christofferson DE, Li Y, Yuan J . Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol 2014; 76: 129–150.
Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV . TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 1996; 4: 387–396.
Hsu H, Shu HB, Pan MG, Goeddel DV . TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 1996; 84: 299–308.
Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, Schmukle AC et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (tnf) to efficiently activate nf-{kappa}b and to prevent tnf-induced apoptosis. J Biol Chem 2009; 2284: 35906–35915.
Ting AT, Pimentel-Muinos FX, Seed B . RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J 1996; 15: 6189–6196.
Gentle IE, Wong WW, Evans JM, Bankovacki A, Cook WD, Khan NR et al. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8. J Biol Chem 2011; 286: 13282–13291.
Lin Y, Devin A, Rodriguez Y, Liu ZG . Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev 1999; 13: 2514–2526.
Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 2011; 471: 591–596.
Lee TH, Shank J, Cusson N, Kelliher MA . The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem 2004; 279: 33185–33191.
Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P . Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol 2014; 15: 135–147.
Micheau O, Tschopp J . Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003; 114: 181–190.
Wang CY, Mayo MW, Baldwin AS Jr . TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science 1996; 274: 784–787.
Chen ZJ . Ubiquitination in signaling to and activation of IKK. Immunol Rev 2012; 246: 95–106.
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123.
He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111.
Christofferson DE, Yuan J . Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol 2010; 22: 263–268.
Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321.
Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 2008; 30: 689–700.
Gaither A, Porter D, Yao Y, Borawski J, Yang G, Donovan J et al. A Smac mimetic rescue screen reveals roles for inhibitor of apoptosis proteins in tumor necrosis factor-alpha signaling. Cancer Res 2007; 67: 11493–11498.
Darding M, Feltham R, Tenev T, Bianchi K, Benetatos C, Silke J et al. Molecular determinants of Smac mimetic induced degradation of cIAP1 and cIAP2. Cell Death Differ 2011; 18: 1376–1386.
Christofferson DE, Li Y, Hitomi J, Zhou W, Upperman C, Zhu H et al. A novel role for RIP1 kinase in mediating TNFalpha production. Cell Death Dis 2012; 3: e320.
**e T, Peng W, Liu Y, Yan C, Maki J, Degterev A et al. Structural basis of RIP1 inhibition by necrostatins. Structure 2013; 21: 493–499.
Berger SB, Kasparcova V, Hoffman S, Swift B, Dare L, Schaeffer M et al. Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol 2014; 192: 5476–5480.
Polykratis A, Hermance N, Zelic M, Roderick J, Kim C, Van TM et al. Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J Immunol 2014; 193: 1539–1543.
Rickard JA, O'Donnell JA, Evans JM, Lalaoui N, Poh AR, Rogers T et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 2014; 157: 1175–1188.
Dillon CP, Weinlich R, Rodriguez DA, Cripps JG, Quarato G, Gurung P et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 2014; 157: 1189–1202.
Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P et al. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 1998; 8: 297–303.
Zhang X, Fan C, Zhang H, Zhao Q, Liu Y, Xu C et al. MLKL and FADD are critical for suppressing progressive lymphoproliferative disease and activating the NLRP3 inflammasome. Cell Rep 2016; 16: 3247–3259.
Alvarez-Diaz S, Dillon CP, Lalaoui N, Tanzer MC, Rodriguez DA, Lin A et al. The pseudokinase MLKL and the kinase RIPK3 have distinct roles in autoimmune disease caused by loss of death-receptor-induced apoptosis. Immunity 2016; 45: 513–526.
Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J et al. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 2011; 471: 373–376.
Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372.
Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367.
Kaiser WJ, Daley-Bauer LP, Thapa RJ, Mandal P, Berger SB, Huang C et al. RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc Natl Acad Sci USA 2014; 111: 7753–7758.
Wu XN, Yang ZH, Wang XK, Zhang Y, Wan H, Song Y et al. Distinct roles of RIP1-RIP3 hetero- and RIP3-RIP3 homo-interaction in mediating necroptosis. Cell Death Differ 2014; 21: 1709–1720.
Rebsamen M, Heinz LX, Meylan E, Michallet MC, Schroder K, Hofmann K et al. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep 2009; 10: 916–922.
de Almagro MC, Vucic D . Necroptosis: pathway diversity and characteristics. Semin Cell Dev Biol 2015; 39: 56–62.
Zhang Y, Su SS, Zhao S, Yang Z, Zhong CQ, Chen X et al. RIP1 autophosphorylation is promoted by mitochondrial ROS and is essential for RIP3 recruitment into necrosome. Nat Commun 2017; 8: 14329.
Galluzzi L, Vanden Berghe T, Vanlangenakker N, Buettner S, Eisenberg T, Vandenabeele P et al. Programmed necrosis from molecules to health and disease. Int Rev Cell Mol Biol 2011; 289: 1–35.
Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012; 150: 339–350.
Zhao XM, Chen Z, Zhao JB, Zhang PP, Pu YF, Jiang SH et al. Hsp90 modulates the stability of MLKL and is required for TNF-induced necroptosis. Cell Death Dis 2016; 7: e2089.
Yoon S, Bogdanov K, Kovalenko A, Wallach D . Necroptosis is preceded by nuclear translocation of the signaling proteins that induce it. Cell Death Differ 2016; 23: 253–260.
Wu J, Huang Z, Ren J, Zhang Z, He P, Li Y et al. Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res 2013; 23: 994–1006.
Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 2011; 477: 330–334.
Fan C, Pu W, Wu X, Zhang X, He L, Zhou B et al. Lack of FADD in Tie-2 expressing cells causes RIPK3-mediated embryonic lethality. Cell Death Dis 2016; 7: e2351.
Kang TB, Ben-Moshe T, Varfolomeev EE, Pewzner-Jung Y, Yogev N, Jurewicz A et al. Caspase-8 serves both apoptotic and nonapoptotic roles. J Immunol 2004; 173: 2976–2984.
Shutinoski B, Alturki NA, Rijal D, Bertin J, Gough PJ, Schlossmacher MG et al. K45A mutation of RIPK1 results in poor necroptosis and cytokine signaling in macrophages, which impacts inflammatory responses in vivo. Cell Death Differ 2016; 23: 1628–1637.
Linkermann A, Brasen JH, Himmerkus N, Liu S, Huber TB, Kunzendorf U et al. Rip1 (receptor-interacting protein kinase 1) mediates necroptosis and contributes to renal ischemia/reperfusion injury. Kidney Int 2012; 81: 751–761.
Newton K, Dugger DL, Maltzman A, Greve JM, Hedehus M, Martin-McNulty B et al. RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ 2016; 23: 1565–1576.
Acknowledgements
We thank Dr **aodong Wang (National Institute of Biological Sciences, Bei**g, China) for providing Ripk3−/− mice and Dr. Jianke Zhang (Thomas Jefferson University, Philadelphia, PA, USA) for providing Fadd+/− mice. We also thank Dr Yu Sun (David Geffen School of Medicine, UCLA, USA) for insightful discussions and critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31571426) and the Ministry of Science and Technology of the People’s Republic of China (2016YFC1304900, 2016YFA0500100). HBZ was supported by Thousand Young Talents Program of the Chinese government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by J. Silke
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, Y., Fan, C., Zhang, Y. et al. RIP1 kinase activity-dependent roles in embryonic development of Fadd-deficient mice. Cell Death Differ 24, 1459–1469 (2017). https://doi.org/10.1038/cdd.2017.78
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2017.78
- Springer Nature Limited
This article is cited by
-
Age-related noncanonical TRMT6–TRMT61A signaling impairs hematopoietic stem cells
Nature Aging (2024)
-
Ubiquitin-binding domain in ABIN1 is critical for regulating cell death and inflammation during development
Cell Death & Differentiation (2022)
-
RIP1 kinase activity promotes steatohepatitis through mediating cell death and inflammation in macrophages
Cell Death & Differentiation (2021)
-
Catalytically inactive RIP1 and RIP3 deficiency protect against acute ischemic stroke by inhibiting necroptosis and neuroinflammation
Cell Death & Disease (2020)
-
Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases
Nature Reviews Neuroscience (2019)