Abstract
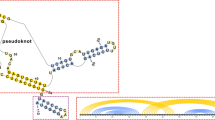
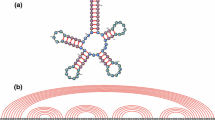
Consider a random word \(X^n=(X_1,\ldots ,X_n)\) in an alphabet consisting of 4 letters, with the letters viewed either as A, U, G and C (i.e., nucleotides in an RNA sequence) or \(\alpha \), \(\bar{\alpha }\), \(\beta \) and \(\bar{\beta }\) (i.e., generators of the free group \(\langle \alpha ,\beta \rangle \) and their inverses). We show that the expected fraction \(\rho (n)\) of unpaired bases in an optimal RNA secondary structure (with only Watson-Crick bonds and no pseudo-knots) converges to a constant \(\lambda _2\) with \(0<\lambda _2<1\) as \(n\rightarrow \infty \). Thus, a positive proportion of the bases of a random RNA string do not form hydrogen bonds. We do not know the exact value of \(\lambda _2\), but we derive upper and lower bounds for it. In terms of free groups, \(\rho (n)\) is the ratio of the length of the shortest word representing X in the generating set consisting of conjugates of generators and their inverses to the word length of X with respect to the standard generators and their inverses. Thus for a typical word the word length in the (infinite) generating set consisting of the conjugates of standard generators grows linearly with the word length in the standard generators. In fact, we show that a similar result holds for all non-abelian finitely generated free groups \(\langle \alpha _1,\dots ,\alpha _k\rangle \), \(k\ge 2\).
Similar content being viewed by others
References
Gadgil, S. Watson-Crick pairing, the Heisenberg group and Milnor invariants, J. Math. Biol. 59, 123 (2009).
Gadgil, S. Conjugacy invariant pseudo-norms, representability and RNA secondary structures, Indian J Pure Appl Math 42, 225 (2011).
Gesteland, R. F. and Cech T. R. and Atkins, J. F. The RNA world: the nature of modern RNA suggests a prebiotic RNA world, Cold Spring Harbor Laboratory Press, (1993).
Kesten, H. Symmetric Random Walks on Groups, Transactions of the American Mathematical Society 92, 336–354.
Ledoux, M. The concentration of measure phenomenon, Mathematical Surveys and Monographs, 89, American Mathematical Society, Providence, RI, 2001.
Steele, J. M. Probability theory and combinatorial optimization, CBMS-NSF Regional Conference Series in Applied Mathematics, 69, Society for Industrial and Applied Mathematics (SIAM), Philadelphia, PA, (1997).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Rajeeva Karandikar.
The first author is partially supported by the SERB-EMR grant EMR/2016/006049. The second author is partially supported by the SERB-MATRICS grant MTR2017/000292. Both authors are partially supported by the UGC centre for advanced studies.
Rights and permissions
About this article
Cite this article
Gadgil, S., Krishnapur, M. Random words in free groups, non-crossing matchings and RNA secondary structures. Indian J Pure Appl Math 54, 146–158 (2023). https://doi.org/10.1007/s13226-022-00240-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13226-022-00240-x