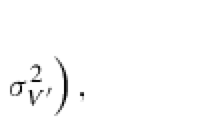Abstract
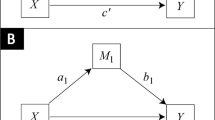
Causal mediation analysis seeks to determine whether an independent variable affects a response variable directly or whether it does so indirectly, by way of a mediator. The existing statistical tests to determine the existence of an indirect effect are overly conservative or have inflated type I error. In this article, we consider the principle of intersection–union tests and a method called the S-test. This method increases power but is not appropriate for statistical tests as small significance levels may cause the test to reject a null hypothesis, but larger significance levels will not reject the same hypothesis. We propose two new methods that provide increased power over existing methods while controlling type I error. We demonstrate through extensive simulation that the S-test and proposed methods control type I error and increase power over existing methods, and that while the proposed methods do not have the same problems, they provide similar power to the S-test. Finally, we provide an application to a large proteomic study.



Similar content being viewed by others
Data availability
The data are not publicly available due to privacy or ethical restrictions.
References
Alwin DF, Hauser RM (1975) The decomposition of effects in path analysis. Am Soc Rev 40(1):37–47. https://doi.org/10.2307/2094445
Aref H, Refaat S (2014) CRP evaluation in non-small cell lung cancer. Egypt J Chest Dis Tuberc 63(3):717–722. https://doi.org/10.1016/j.ejcdt.2014.02.003
Barfield R, Shen J, Just AC, Vokonas PS, Schwartz J, Baccarelli AA, VanderWeele TJ, Lin X (2017) Testing for the indirect effect under the null for genome-wide mediation analyses. Genet Epidemiol 41(8):824–833. https://doi.org/10.1002/gepi.22084
Bellavia A, James-Todd T, Williams PL (2019) Approaches for incorporating environmental mixtures as mediators in mediation analysis. Environ Int 123:368–374. https://doi.org/10.1016/j.envint.2018.12.024
Berger RL (1997) Likelihood ratio tests and intersection-union tests. In: Panchapakesan S, Balakrishnan N (eds) Advances in statistical decision theory and applications. Birkhäuser, Boston, pp 225–237. https://doi.org/10.1007/978-1-4612-2308-5_15
Biesanz JC, Falk CF, Savalei V (2010) Assessing mediational models: testing and interval estimation for indirect effects. Multivar Behav Res 45(4):661–701. https://doi.org/10.1080/00273171.2010.498292
Cardenas A, Lutz SM, Everson TM, Perron P, Bouchard L, Hivert MF (2019) Mediation by placental DNA methylation of the association of prenatal maternal smoking and birth weight. Am J Epidemiol 188(11):1878–1886. https://doi.org/10.1093/aje/kwz184
Cho SH, Huang YT (2019) Mediation analysis with causally ordered mediators using Cox proportional hazards model. Stat Med 38(9):1566–1581. https://doi.org/10.1002/sim.8058
Cohen J, Cohen P (1983) Applied multiple regression/correlation analysis for the behavioral sciences. Taylor & Francis Group, New York
Couper D, LaVange LM, Han M, Barr RG, Bleecker E, Hoffman EA, Kanner R, Kleerup E, Martinez FJ, Woodruff PG (2014) Design of the subpopulations and intermediate outcomes in COPD study (SPIROMICS). Thorax 69(5):492–495. https://doi.org/10.1136/thoraxjnl-2013-203897
Dai JY, Stanford JL, LeBlanc M (2020) A multiple-testing procedure for high-dimensional mediation hypotheses. J Am Stat Assoc. https://doi.org/10.1080/01621459.2020.1765785
Derkach A, Moore S, Boca S, Sampson J (2020) Group testing in mediation analysis. Stat Med. https://doi.org/10.1002/sim.8546
Derkach A, Pfeiffer R, Chen T, Sampson J (2019) High dimensional mediation analysis with latent variables. Biometrics. https://doi.org/10.1111/biom.13053
Foreman MG, Kong X, DeMeo DL, Pillai SG, Hersh CP, Bakke P, Gulsvik A, Lomas DA, Litonjua AA, Shapiro SD, Tal-Singer R, Silverman EK (2011) Polymorphisms in surfactant protein-D are associated with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 44(3):316–322. https://doi.org/10.1165/rcmb.2009-0360OC
Huang YT (2018) Joint significance tests for mediation effects of socioeconomic adversity on adiposity via epigenetics. Ann Appl Stat 12(3):1535–1557. https://doi.org/10.1214/17-AOAS1120
Huang YT (2019) Genome-wide analyses of sparse mediation effects under composite null hypotheses. Ann Appl Stat 13:60–84
Huang YT, Pan WC (2016) Hypothesis test of mediation effect in causal mediation model with high-dimensional continuous mediators. Biometrics 72(2):402–413. https://doi.org/10.1111/biom.12421
Huang YT, VanderWeele TJ, Lin X (2014) Joint analysis of SNP and gene expression data in genetic association studies of complex diseases. Ann Appl Stat 8(1):1–24. https://doi.org/10.1214/13-AOAS690.JOINT
Hutton J, Fatima T, Major TJ, Topless R, Stamp LK, Merriman TR, Dalbeth N (2018) Mediation analysis to understand genetic relationships between habitual coffee intake and gout. Arth Res Ther 20(1):135. https://doi.org/10.1186/s13075-018-1629-5
Imai K, Keele L, Yamamoto T (2010) Identification, inference and sensitivity analysis for causal mediation effects. Stat Sci 25(1):51–71. https://doi.org/10.1214/10-STS321. ar**v:1011.1079
Kisbu-Sakarya Y, MacKinnon DP, Miočević M (2014) The distribution of the product explains normal theory mediation confidence interval estimation. Multivar Behav Res 49(3):261–268. https://doi.org/10.1080/00273171.2014.903162
Lin DY, Zeng D, Couper D (2020) A general framework for integrative analysis of incomplete multiomics data. Genet Epidemiol 44(7):646–664. https://doi.org/10.1002/gepi.22328
MacKinnon DP, Fairchild AJ, Fritz MS (2007) Mediation analysis. Annu Rev Psychol 58:593–614. https://doi.org/10.1146/annurev.psych.58.110405.085542
MacKinnon DP, Lockwood CM, Hoffman JM, West SG, Sheets V (2002) A comparison of methods to test mediation and other intervening variable effects. Psychol Methods 7(1):83–104. https://doi.org/10.1037//1082-989X.7.1.83. ar**v:0412114v1 [ar**v:cs]
MacKinnon DP, Lockwood CM, Williams J (2004) Confidence limits for the indirect effect: Distribution of the product and resampling methods. Multivar Behav Res 39(1):99–128. https://doi.org/10.1207/s15327906mbr3901_4
Nandy D, Sharma N, Senapati S (2019) Systematic review and meta-analysis confirms significant contribution of surfactant protein D in chronic obstructive pulmonary disease. Front Genet 10:1–7. https://doi.org/10.3389/fgene.2019.00339
...Obeidat M, Li X, Burgess S, Zhou G, Fishbane N, Hansel NN, Bossé Y, Joubert P, Hao K, Nickle DC, van den Berge M, Timens W, Cho MH, Hobbs BD, de Jong K, Boezen M, Hung RJ, Rafaels N, Mathias R, Ruczinski I, Beaty TH, Barnes KC, Paré PD, Sin DD (2017) Surfactant protein D is a causal risk factor for COPD: results of Mendelian randomisation. Eur Respir J 50(5):1–11. https://doi.org/10.1183/13993003.00657-2017
...O’Neal WK, Anderson W, Basta PV, Carretta EE, Doerschuk CM, Barr RG, Bleecker ER, Christenson SA, Curtis JL, Han MK, O’Neal WK, Anderson W, Basta PV, Carretta EE, Doerschuk CM, Barr RG, Bleecker ER, Christenson SA, Curtis JL, Han MK, Hansel NN, Kanner RE, Kleerup EC, Martinez FJ, Miller BE, Peters SP, Rennard SI, Scholand MB, Tal-Singer R, Woodruff PG, Couper DJ, Davis SM (2014) Comparison of serum, EDTA plasma and P100 plasma for luminex-based biomarker multiplex assays in patients with chronic obstructive pulmonary disease in the SPIROMICS study. J Transl Med 12(1):1–9. https://doi.org/10.1186/1479-5876-12-9
...Raulerson CK, Ko A, Kidd JC, Currin KW, Brotman SM, Cannon ME, Wu Y, Spracklen CN, Jackson AU, Stringham HM, Welch RP, Fuchsberger C, Locke AE, Narisu N, Lusis AJ, Civelek M, Furey TS, Kuusisto J, Collins FS, Boehnke M, Scott LJ, Lin DY, Love MI, Laakso M, Pajukanta P, Mohlke KL (2019) Adipose tissue gene expression associations reveal hundreds of candidate genes for cardiometabolic traits. Am J Hum Genet 105(4):773–787. https://doi.org/10.1016/j.ajhg.2019.09.001
Richiardi L, Bellocco R, Zugna D (2013) Mediation analysis in epidemiology: methods, interpretation and bias. Int J Epidemiol 42(5):1511–1519. https://doi.org/10.1093/ije/dyt127
Sobel ME (1982) Asymptotic confidence intervals for indirect effects in structural equation models. Soc Methodol 13:290–312. https://doi.org/10.2307/270723
Sun, W., K. Kechris, S. Jacobson, M.B. Drummond, G.A. Hawkins, J. Yang, T.h. Chen, P.M. Quibrera, W. Anderson, R.G. Barr, P.V. Basta, E.R. Bleecker, T. Beaty, R. Casaburi, P. Castaldi, M.H. Cho, A. Comellas, J.D. Crapo, G. Criner, D. Demeo, S.A. Christenson, D.J. Couper, J.L. Curtis, C.M. Doerschuk, C.M. Freeman, N.A. Gouskova, M.K. Han, N.A. Hanania, N.N. Hansel, C.P. Hersh, E.A. Hoffman, R.J. Kaner, R.E. Kanner, E.C. Kleerup, S. Lutz, F.J. Martinez, D.A. Meyers, S.P. Peters, E.A. Regan, S.I. Rennard, M.B. Scholand, E.K. Silverman, P.G. Woodruff, W.K. O’Neal, R.P. Bowler, S.R. Group, and C.I. Investigators (2016) Common genetic polymorphisms influence blood biomarker measurements in COPD. PLOS Genet 12(8):e1006011. https://doi.org/10.1371/journal.pgen.1006011
Vanderweele TJ (2011) Controlled Direct and Mediated Effects: Definition, Identification and Bounds. Scand J Stat 38(3):551–563. https://doi.org/10.1111/j.1467-9469.2010.00722.x
Vanderweele TJ, Vansteelandt S (2009) Conceptual issues concerning mediation, interventions and composition. Stat Interface 2(4):457–468. https://doi.org/10.4310/SII.2009.v2.n4.a7
VanderWeele TJ, Vansteelandt S (2013) Mediation analysis with multiple mediators. Epidemiol Methods 2(1):95–115. https://doi.org/10.1515/em-2012-0010.ar**v:NIHMS150003
Wang K (2018) Understanding power anomalies in mediation analysis. Psychometrika 83(2):387–406. https://doi.org/10.1007/s11336-017-9598-1
Zhang W, Fan J, Chen Q, Lei C, Qiao B, Liu Q (2018) SPP1 and AGER as potential prognostic biomarkers for lung adenocarcinoma. Oncol Lett 15(5):7028–7036. https://doi.org/10.3892/ol.2018.8235
Zhong W, Darville T, Zheng X, Fine J, Li Y (2020) Biometrics, Generalized multi-SNP mediation intersection-union test. https://doi.org/10.1111/biom.13418
Zhong W, Spracklen CN, Mohlke KL, Zheng X, Fine J, Li Y (2019) Multi-SNP mediation intersection-union test. Bioinformatics 35(22):4724–4729. https://doi.org/10.1093/bioinformatics/btz285
Acknowledgements
We thank Dr. Chris Shendhal for processing the SPIROMICS data.
Funding
Research reported in this publication was supported by the National Institutes Of Health under Award Number R01HG009974.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no potential competing of interests.
Appendices
Appendix A
Potential Inflation of the Type I Error for the PS-Test
Due to the symmetric nature of the rejection region, the potential inflation of the Type I error for the PS-test is the same when \(\gamma = 0\) and \(\beta \ne 0\) versus when \(\beta = 0\) and \(\gamma \ne 0\). Thus, we assume that \(\gamma = 0\) and \(\beta \ne 0\), in which case \(U_{\gamma }\) is standard uniform. The probability that \((U_{\beta }, U_{\gamma })\) lies within the rejection region depends on the value of \(\alpha\) (just like the original S-test) and on the value of \(U_{\beta }\) (unlike the original S-test). Let \(g(u_{\beta })\) denote the probability that \(H_0\) is rejected for the chosen \(\alpha\) when \(U_{\beta }=u_{\beta }\), and let \(f_{U_{\beta }}(u_\beta )\) denote the density function of \(U_{\beta }\). The probability of making a type I error at the significance level \(\alpha\) equals \(\int _0^1 g(x) f_{U_{\beta }}(x) dx\). We can determine the noncentrality parameter of \(T_{\beta }\) that causes the largest type I error for any value of \(\alpha\) and then determine the maximum inflation of the type I error over all possible values of \(\alpha\).
We use numerical integration to calculate the type I error. We consider both small-sample and asymptotic scenarios, using a noncentral t-distribution with five degrees of freedom and a normal distribution with mean equal to the noncentrality parameter and unit variance. In the small-sample scenario, the maximum possible type I error rate occurs when \(\alpha \approx 0.002\) and is approximately 1.0001 times \(\alpha\). In the asymptotic case, the maximum type I error occurs when \(\alpha \approx 0.028\) and is approximately 1.0084 times \(\alpha\). In each case, the increase in the type I error for the PS-test is less than 1% of \(\alpha\), and more common choices of \(\alpha\) have even lower inflation of the type I error.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kidd, J., Lin, DY. Improving the Power to Detect Indirect Effects in Mediation Analysis. Stat Biosci 16, 129–141 (2024). https://doi.org/10.1007/s12561-023-09386-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12561-023-09386-6