Abstract
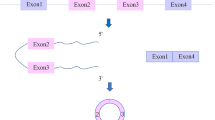
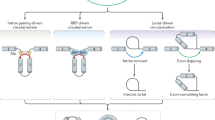
Circular RNAs (circRNAs) are single-strand covalently closed circular noncoding RNAs that are endogenous transcripts generated from linear precursor mRNA through a backsplicing mechanism. With the development of high-throughput sequencing technology, a number of circRNAs have been identified and proved to play key roles in various pathophysiological processes, such as metabolic diseases, cancers, and cardiovascular diseases. An increasing number of studies have shown that circRNAs are widely expressed in cardiac tissues and play important roles in the development of multiple cardiovascular diseases. Here, we review the current understanding of circRNA biogenesis and functions and the roles of circRNAs in cardiovascular diseases. We also highlight the molecular mechanisms underlying the role of circRNAs in the pathogenesis of cardiovascular diseases. A better understanding of the biological function of circRNAs in cardiovascular diseases will be helpful for the development of effective biomarkers for the diagnosis and treatment of these diseases.



Similar content being viewed by others
References
Han, B., Chao, J., & Yao, H. (2018). Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacology & Therapeutics, 187, 31–44. https://doi.org/10.1016/j.pharmthera.2018.01.010.
Fischer, J. W., & Leung, A. K. (2017). CircRNAs: a regulator of cellular stress. Critical Reviews in Biochemistry and Molecular Biology, 52(2), 220–233. https://doi.org/10.1080/10409238.2016.1276882.
Jeck, W. R., & Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nature Biotechnology, 32(5), 453–461. https://doi.org/10.1038/nbt.2890.
Abdelmohsen, K., Panda, A. C., Munk, R., Grammatikakis, I., Dudekula, D. B., De, S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biology, 14(3), 361–369. https://doi.org/10.1080/15476286.2017.1279788.
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell, 56(1), 55–66. https://doi.org/10.1016/j.molcel.2014.08.019.
Shan, K., Liu, C., Liu, B. H., Chen, X., Dong, R., Liu, X., et al. (2017). Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation, 136(17), 1629–1642. https://doi.org/10.1161/circulationaha.117.029004.
Boeckel, J. N., Jae, N., Heumuller, A. W., Chen, W., Boon, R. A., Stellos, K., et al. (2015). Identification and characterization of hypoxia-regulated endothelial circular RNA. Circulation Research, 117(10), 884–890. https://doi.org/10.1161/circresaha.115.306319.
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., & Yang, B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research, 44(6), 2846–2858. https://doi.org/10.1093/nar/gkw027.
Jordan, M. C., Henderson, S. A., Han, T., Fishbein, M. C., Philipson, K. D., & Roos, K. P. (2010). Myocardial function with reduced expression of the sodium-calcium exchanger. Journal of Cardiac Failure, 16(9), 786–796. https://doi.org/10.1016/j.cardfail.2010.03.012.
Fang, Y., Wang, X., Li, W., Han, J., **, J., Su, F., et al. (2018). Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. International Journal of Molecular Medicine, 42(4), 1865–1874. https://doi.org/10.3892/ijmm.2018.3783.
Klinge, C. (2018). Non-coding RNAs in breast cancer: intracellular and intercellular communication. Non-Coding RNA, 4(4), 40. https://doi.org/10.3390/ncrna4040040.
Shang, Q., Yang, Z., Jia, R., & Ge, S. (2019). The novel roles of circRNAs in human cancer. Molecular Cancer, 18(1), 6. https://doi.org/10.1186/s12943-018-0934-6.
Wang, K., Long, B., Liu, F., Wang, J. X., Liu, C. Y., Zhao, B., et al. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European Heart Journal, 37(33), 2602–2611. https://doi.org/10.1093/eurheartj/ehv713.
Townsend, N., Nichols, M., Scarborough, P., & Rayner, M. (2015). Cardiovascular disease in Europe--epidemiological update 2015. European Heart Journal, 36(40), 2696–2705. https://doi.org/10.1093/eurheartj/ehv428.
Townsend, N., Wilson, L., Bhatnagar, P., Wickramasinghe, K., Rayner, M., & Nichols, M. (2016). Cardiovascular disease in Europe: epidemiological update 2016. European Heart Journal, 37(42), 3232–3245. https://doi.org/10.1093/eurheartj/ehw334.
Li, M., Ding, W., Sun, T., Tariq, M. A., Xu, T., Li, P., et al. (2018). Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. The FEBS Journal, 285(2), 220–232. https://doi.org/10.1111/febs.14191.
Wang, H., Yang, J., Yang, J., Fan, Z., & Yang, C. (2016). Circular RNAs: novel rising stars in cardiovascular disease research. International Journal of Cardiology, 202, 726–727. https://doi.org/10.1016/j.ijcard.2015.10.051.
Devaux, Y., Creemers, E. E., Boon, R. A., Werfel, S., Thum, T., Engelhardt, S., et al. (2017). Circular RNAs in heart failure. European Journal of Heart Failure, 19(6), 701–709. https://doi.org/10.1002/ejhf.801.
Fan, X., Weng, X., Zhao, Y., Chen, W., Gan, T., & Xu, D. (2017). Circular RNAs in cardiovascular disease: an overview. BioMed Research International, 2017, 5135781. https://doi.org/10.1155/2017/5135781.
Memczak, S., Papavasileiou, P., Peters, O., & Rajewsky, N. (2015). Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One, 10(10), e0141214. https://doi.org/10.1371/journal.pone.0141214.
Greene, J., Baird, A. M., Brady, L., Lim, M., Gray, S. G., McDermott, R., et al. (2017). Circular RNAs: biogenesis, function and role in human diseases. Frontiers in Molecular Biosciences, 4, 38. https://doi.org/10.3389/fmolb.2017.00038.
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., & Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proceedings of the National Academy of Sciences of the United States of America, 73(11), 3852–3856.
Hsu, M. T., & Coca-Prados, M. (1979). Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature, 280(5720), 339–340.
Cocquerelle, C., Daubersies, P., Majerus, M. A., Kerckaert, J. P., & Bailleul, B. (1992). Splicing with inverted order of exons occurs proximal to large introns. The EMBO Journal, 11(3), 1095–1098.
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled exons. Cell, 64(3), 607–613. https://doi.org/10.1016/0092-8674(91)90244-S.
Pasman, Z., Been, M. D., & Garcia-Blanco, M. A. (1996). Exon circularization in mammalian nuclear extracts. RNA, 2(6), 603–610.
Capel, B., Swain, A., Nicolis, S., Hacker, A., Walter, M., Koopman, P., et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell, 73(5), 1019–1030. https://doi.org/10.1016/0092-8674(93)90279-Y.
Surono, A., Takeshima, Y., Wibawa, T., Ikezawa, M., Nonaka, I., & Matsuo, M. (1999). Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Human Molecular Genetics, 8(3), 493–500.
Li, X. F., & Lytton, J. (1999). A circularized sodium-calcium exchanger exon 2 transcript. The Journal of Biological Chemistry, 274(12), 8153–8160. https://doi.org/10.1074/jbc.274.12.8153.
Houseley, J. M., Garcia-Casado, Z., Pascual, M., Paricio, N., O’Dell, K. M., Monckton, D. G., et al. (2006). Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. The Journal of Heredity, 97(3), 253–260. https://doi.org/10.1093/jhered/esj037.
Cocquerelle, C., Mascrez, B., Hetuin, D., & Bailleul, B. (1993). Mis-splicing yields circular RNA molecules. The FASEB Journal, 7(1), 155–160.
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., & Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One, 7(2), e30733. https://doi.org/10.1371/journal.pone.0030733.
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L., & Brown, P. O. (2013). Cell-type specific features of circular RNA expression. PLoS Genetics, 9(9), e1003777. https://doi.org/10.1371/journal.pgen.1003777.
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19(2), 141–157. https://doi.org/10.1261/rna.035667.112.
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. https://doi.org/10.1038/nature11928.
Guo, J. U., Agarwal, V., Guo, H., & Bartel, D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biology, 15(7), 409. https://doi.org/10.1186/s13059-014-0409-z.
Wang, Y., & Wang, Z. (2015). Efficient backsplicing produces translatable circular mRNAs. RNA, 21(2), 172–179. https://doi.org/10.1261/rna.048272.114.
Westholm, J. O., Miura, P., Olson, S., Shenker, S., Joseph, B., Sanfilippo, P., et al. (2014). Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Reports, 9(5), 1966–1980. https://doi.org/10.1016/j.celrep.2014.10.062.
Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., & Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell, 159(1), 134–147. https://doi.org/10.1016/j.cell.2014.09.001.
Veno, M. T., Hansen, T. B., Veno, S. T., Clausen, B. H., Grebing, M., Finsen, B., et al. (2015). Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biology, 16, 245. https://doi.org/10.1186/s13059-015-0801-3.
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495(7441), 384–388. https://doi.org/10.1038/nature11993.
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol Cell, 66(1), 9–21.e27. https://doi.org/10.1016/j.molcel.2017.02.021.
Burset, M., Seledtsov, I. A., & Solovyev, V. V. (2000). Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Research, 28(21), 4364–4375. https://doi.org/10.1093/nar/28.21.4364.
Altesha, M. A., Ni, T., Khan, A., Liu, K., & Zheng, X. (2019). Circular RNA in cardiovascular disease. Journal of Cellular Physiology, 234(5), 5588–5600. https://doi.org/10.1002/jcp.27384.
Li, X., Yang, L., & Chen, L. L. (2018). The biogenesis, functions, and challenges of circular RNAs. Molecular Cell, 71(3), 428–442. https://doi.org/10.1016/j.molcel.2018.06.034.
Chen, L. L., & Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biology, 12(4), 381–388. https://doi.org/10.1080/15476286.2015.1020271.
Chen, W., & Schuman, E. (2016). Circular RNAs in brain and other tissues: a functional enigma. Trends in Neurosciences, 39(9), 597–604. https://doi.org/10.1016/j.tins.2016.06.006.
Ebbesen, K. K., Kjems, J., & Hansen, T. B. (2016). Circular RNAs: identification, biogenesis and function. Biochimica et Biophysica Acta, 1859(1), 163–168. https://doi.org/10.1016/j.bbagrm.2015.07.007.
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology, 22(3), 256–264. https://doi.org/10.1038/nsmb.2959.
Zhang, Y., Zhang, X. O., Chen, T., **ang, J. F., Yin, Q. F., **ng, Y. H., et al. (2013). Circular intronic long noncoding RNAs. Molecular Cell, 51(6), 792–806. https://doi.org/10.1016/j.molcel.2013.08.017.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., & Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146(3), 353–358. https://doi.org/10.1016/j.cell.2011.07.014.
Piwecka, M., Glazar, P., Hernandez-Miranda, L. R., Memczak, S., Wolf, S. A., Rybak-Wolf, A., et al. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science, 357(6357), eaam8526. https://doi.org/10.1126/science.aam8526.
Holdt, L. M., Stahringer, A., Sass, K., Pichler, G., Kulak, N. A., Wilfert, W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nature Communications, 7, 12429. https://doi.org/10.1038/ncomms12429.
Chen, C. Y., & Sarnow, P. (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science, 268(5209), 415–417.
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell, 66(1), 22–37.e29. https://doi.org/10.1016/j.molcel.2017.02.017.
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Research, 27(5), 626–641. https://doi.org/10.1038/cr.2017.31.
Conn, V. M., Hugouvieux, V., Nayak, A., Conos, S. A., Capovilla, G., Cildir, G., et al. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. [Letter]. Nat Plants, 3, 17053. https://doi.org/10.1038/nplants.2017.53.
Werfel, S., Nothjunge, S., Schwarzmayr, T., Strom, T. M., Meitinger, T., & Engelhardt, S. (2016). Characterization of circular RNAs in human, mouse and rat hearts. Journal of Molecular and Cellular Cardiology, 98, 103–107. https://doi.org/10.1016/j.yjmcc.2016.07.007.
Jakobi, T., Czaja-Hasse, L. F., Reinhardt, R., & Dieterich, C. (2016). Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics, Proteomics & Bioinformatics, 14(4), 216–223. https://doi.org/10.1016/j.gpb.2016.02.003.
Tan, W. L., Lim, B. T., Anene-Nzelu, C. G., Ackers-Johnson, M., Dashi, A., See, K., et al. (2017). A landscape of circular RNA expression in the human heart. Cardiovascular Research, 113(3), 298–309. https://doi.org/10.1093/cvr/cvw250.
Broadbent, H. M., Peden, J. F., Lorkowski, S., Goel, A., Ongen, H., Green, F., et al. (2008). Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Human Molecular Genetics, 17(6), 806–814. https://doi.org/10.1093/hmg/ddm352.
Hall Ignacio, F., Climent, M., Quintavalle, M., Farina Floriana, M., Schorn, T., Zani, S., et al. (2019). Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circulation Research, 124(4), 498–510. https://doi.org/10.1161/CIRCRESAHA.118.314240.
Yang, L., Yang, F., Zhao, H., Wang, M., & Zhang, Y. (2019). Circular RNA circCHFR facilitates the proliferation and migration of vascular smooth muscle via miR-370/FOXO1/cyclin D1 pathway. Molecular Therapy--Nucleic Acids, 16, 434–441. https://doi.org/10.1016/j.omtn.2019.02.028.
Geng, H.-H., Li, R., Su, Y.-M., **ao, J., Pan, M., Cai, X.-X., et al. (2016). The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One, 11(3), e0151753–e0151753. https://doi.org/10.1371/journal.pone.0151753.
Deng, Y.-Y., Zhang, W., She, J., Zhang, L., Chen, T., Zhou, J., et al. (2016). GW27-e1167 circular RNA related to PPARγ function as ceRNA of microRNA in human acute myocardial infarction. Journal of the American College of Cardiology, 68(16, Supplement), C51–C52. https://doi.org/10.1016/j.jacc.2016.07.189.
Li, M., Ding, W., Tariq, M. A., Chang, W., Zhang, X., Xu, W., et al. (2018). A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics, 8(21), 5855–5869. https://doi.org/10.7150/thno.27285.
Wang, K., Gan, T. Y., Li, N., Liu, C. Y., Zhou, L. Y., Gao, J. N., et al. (2017). Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death and Differentiation, 24(6), 1111–1120. https://doi.org/10.1038/cdd.2017.61.
Huang, S., Li, X., Zheng, H., Si, X., Li, B., Wei, G., et al. (2019). Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation, 139(25), 2857–2876. https://doi.org/10.1161/circulationaha.118.038361.
Zhou, L. Y., Zhai, M., Huang, Y., Xu, S., An, T., Wang, Y. H., et al. (2019). The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death and Differentiation, 26(7), 1299–1315. https://doi.org/10.1038/s41418-018-0206-4.
Tang, C. M., Zhang, M., Huang, L., Hu, Z. Q., Zhu, J. N., **ao, Z., et al. (2017). CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Scientific Reports, 7, 40342. https://doi.org/10.1038/srep40342.
Zhou, B., & Yu, J. W. (2017). A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochemical and Biophysical Research Communications, 487(4), 769–775. https://doi.org/10.1016/j.bbrc.2017.04.044.
Zhu, Y., Pan, W., Yang, T., Meng, X., Jiang, Z., Tao, L., et al. (2019). Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR-433. Frontiers in Genetics, 10, 564. https://doi.org/10.3389/fgene.2019.00564.
Khan Mohsin, A. F., Reckman Yolan, J., Aufiero, S., van den Hoogenhof Maarten, M. G., van der Made, I., Beqqali, A., et al. (2016). RBM20 regulates circular RNA production from the titin gene. Circulation Research, 119(9), 996–1003. https://doi.org/10.1161/CIRCRESAHA.116.309568.
Lim, T. B., Aliwarga, E., Luu, T. D. A., Li, Y. P., Ng, S. L., Annadoray, L., et al. (2019). Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovascular Research. https://doi.org/10.1093/cvr/cvz130.
Du, W. W., Yang, W., Chen, Y., Wu, Z. K., Foster, F. S., Yang, Z., et al. (2017). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European Heart Journal, 38(18), 1402–1412. https://doi.org/10.1093/eurheartj/ehw001.
Vausort, M., Salgado-Somoza, A., Zhang, L., Leszek, P., Scholz, M., Teren, A., et al. (2016). Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. Journal of the American College of Cardiology, 68(11), 1247–1248. https://doi.org/10.1016/j.jacc.2016.06.040.
Bazan, H. A., Hatfield, S. A., Brug, A., Brooks, A. J., Lightell, D. J., Jr., & Woods, T. C. (2017). Carotid plaque rupture is accompanied by an increase in the ratio of serum circR-284 to miR-221 levels. Circulation. Cardiovascular Genetics, 10(4), e001720. https://doi.org/10.1161/circgenetics.117.001720.
Wang, L., Shen, C., Wang, Y., Zou, T., Zhu, H., Lu, X., et al. (2019). Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis, 286, 88–96. https://doi.org/10.1016/j.atherosclerosis.2019.05.006.
(2016). Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet, 388(10053), 1459–1544. https://doi.org/10.1016/s0140-6736(16)31012-1.
Biros, E., Cooper, M., Palmer, L. J., Walker, P. J., Norman, P. E., & Golledge, J. (2010). Association of an allele on chromosome 9 and abdominal aortic aneurysm. Atherosclerosis, 212(2), 539–542. https://doi.org/10.1016/j.atherosclerosis.2010.06.015.
Bown, M. J., Braund, P. S., Thompson, J., London, N. J., Samani, N. J., & Sayers, R. D. (2008). Association between the coronary artery disease risk locus on chromosome 9p21.3 and abdominal aortic aneurysm. Circulation. Cardiovascular Genetics, 1(1), 39–42. https://doi.org/10.1161/circgenetics.108.789727.
Ye, S., Willeit, J., Kronenberg, F., Xu, Q., & Kiechl, S. (2008). Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. Journal of the American College of Cardiology, 52(5), 378–384. https://doi.org/10.1016/j.jacc.2007.11.087.
Dietrich, N., Bracken, A. P., Trinh, E., Schjerling, C. K., Koseki, H., Rappsilber, J., et al. (2007). Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. The EMBO Journal, 26(6), 1637–1648. https://doi.org/10.1038/sj.emboj.7601632.
Jacobs, J. J., Kieboom, K., Marino, S., DePinho, R. A., & van Lohuizen, M. (1999). The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature, 397(6715), 164–168. https://doi.org/10.1038/16476.
Swynghedauw, B. (1999). Molecular mechanisms of myocardial remodeling. Physiological Reviews, 79(1), 215–262. https://doi.org/10.1152/physrev.1999.79.1.215.
Zheng, Q., Bao, C., Guo, W., Li, S., Chen, J., Chen, B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications, 7, 11215. https://doi.org/10.1038/ncomms11215.
Dong, Y., Liu, C., Zhao, Y., Ponnusamy, M., Li, P., & Wang, K. (2018). Role of noncoding RNAs in regulation of cardiac cell death and cardiovascular diseases. Cellular and Molecular Life Sciences, 75(2), 291–300. https://doi.org/10.1007/s00018-017-2640-8.
Bayoumi, A. S., Aonuma, T., Teoh, J. P., Tang, Y. L., & Kim, I. M. (2018). Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacologica Sinica, 39(7), 1100–1109. https://doi.org/10.1038/aps.2017.196.
Wu, H. J., Zhang, C. Y., Zhang, S., Chang, M., & Wang, H. Y. (2016). Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cellular Physiology and Biochemistry, 39(1), 205–216. https://doi.org/10.1159/000445617.
Heumüller Andreas, W., & Dimmeler, S. (2019). Circular RNA control of vascular smooth muscle cell functions. Circulation Research, 124(4), 456–458. https://doi.org/10.1161/CIRCRESAHA.118.314521.
Wu, Q. Q., **ao, Y., Yuan, Y., Ma, Z. G., Liao, H. H., Liu, C., et al. (2017). Mechanisms contributing to cardiac remodelling. Clinical Science (London, England), 131(18), 2319–2345. https://doi.org/10.1042/cs20171167.
Steinberg, B. A., Mulpuru, S. K., Fang, J. C., & Gersh, B. J. (2017). Sudden death mechanisms in nonischemic cardiomyopathies: Insights gleaned from clinical implantable cardioverter-defibrillator trials. Heart Rhythm, 14(12), 1839–1848. https://doi.org/10.1016/j.hrthm.2017.09.025.
Kimura, W., ** identifies cycling cardiomyocytes in the adult heart. Nature, 523(7559), 226–230. https://doi.org/10.1038/nature14582.
Gupta, S. K., Garg, A., Bar, C., Chatterjee, S., Foinquinos, A., Milting, H., et al. (2018). Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circulation Research, 122(2), 246–254. https://doi.org/10.1161/circresaha.117.311335.
Koh, W., Pan, W., Gawad, C., Fan, H. C., Kerchner, G. A., Wyss-Coray, T., et al. (2014). Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proceedings of the National Academy of Sciences of the United States of America, 111(20), 7361–7366. https://doi.org/10.1073/pnas.1405528111.
Zhao, Z., Li, X., Gao, C., Jian, D., Hao, P., Rao, L., et al. (2017). Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Scientific Reports, 7, 39918. https://doi.org/10.1038/srep39918.
Funding
This work was funded by the National Natural Science Foundation of China (81741173, 31430041).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Associate Editor Joost Sluijter oversaw the review of this article
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, B., Li, Y., Hu, L. et al. Role of Circular RNAs in the Pathogenesis of Cardiovascular Disease. J. of Cardiovasc. Trans. Res. 13, 572–583 (2020). https://doi.org/10.1007/s12265-019-09912-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-019-09912-2