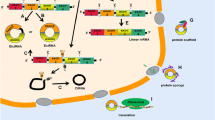
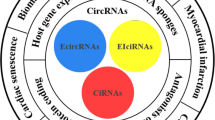
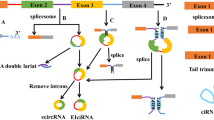
Abstract
Cardiovascular disease (CVD) is one of the vital causes of morbidity and mortality, and the number of deaths from CVD has increased worldwide. Circular RNAs (circRNAs) is a novel type of endogenous noncoding RNA, which can form covalent closed continuous rings and are highly expressed in the eukaryotic transcriptome. In recent years, research on circRNAs have been increasing and the researchers have also become cumulatively aware of the association between circRNAs and CVD. This review highlights the biogenesis and functions of circRNAs and the role in cardiovascular diseases.





Similar content being viewed by others
Abbreviations
- CVD:
-
Cardiovascular disease
- circRNAs:
-
Circular RNAs
- EIciRNAs:
-
Exon-intron circRNAs
- ciRNAs:
-
Circular intronic RNAs
- ciRS-7:
-
Circular RNA sponge for miR-7
- SR:
-
Serine-arginine
- hnRNP:
-
Heterogeneous nuclear ribonucleoprotein
- Pol II:
-
Polymerase II
- snRNPs:
-
Small nuclear ribonucleoproteins
- CDK2:
-
Cyclin-dependent kinase 2
- HUVECs:
-
Human umbilical vein endothelial cells
- cANRIL:
-
Circular antisense noncoding RNA in the INK4 locus
- SNPs:
-
Single nucleotide polymorphisms
- PES1:
-
Pescadillo homologue 1
- TAC:
-
Thoracic aortic constriction
- HRCR:
-
Heart-related circRNA
- IGF-1:
-
Insulin-like growth factors-1
- NRG-1-ICD:
-
Neuregulin-1 intracellular domain
- α-SMA:
-
Smooth muscle α-actin
- EH:
-
Essential hypertension
- TGF:
-
Transforming growth factor
References
AbuQattam A, Serrano-Quílez J, Rodríguez-Navarro S, Gallego J (2018) An exon three-way junction structure modulates splicing and degradation of the SUS1 yeast pre-mRNA. Biochim Biophys Acta Gene Regul Mech 1861:673–686. https://doi.org/10.1016/j.bbagrm.2018.06.009
Adamcova M, Simko F (2018) Multiplex biomarker approach to cardiovascular diseases. Acta Pharmacol Sin 39:1068–1072. https://doi.org/10.1038/aps.2018.29
Akhter R (2018) Circular RNA and Alzheimer’s disease. Adv Exp Med Biol 1087:239–243. https://doi.org/10.1007/978-981-13-1426-1_19
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J, Zhang L (2018) A potential risk factor of essential hypertension in case-control study: circular RNA hsa_circ_0037911. Biochem Biophys Res Commun 498:789–794. https://doi.org/10.1016/j.bbrc.2018.03.059
Bis** E, Wakula P, Poteser M, Heinzel FR (2014) Targeting cardiac hypertrophy: toward a causal heart failure therapy. J Cardiovasc Pharmacol 64:293–305. https://doi.org/10.1097/fjc.0000000000000126
Bitton D, Atkinson S, Rallis C, Smith G, Ellis D, Chen Y, Malecki M, Codlin S, Lemay J, Cotobal C, Bachand F, Marguerat S, Mata J, Bähler J (2015) Widespread exon skip** triggers degradation by nuclear RNA surveillance in fission yeast. Genome Res 25:884–896. https://doi.org/10.1101/gr.185371.114
Bugger H, Abel ED (2014) Molecular mechanisms of diabetic cardiomyopathy. Diabetologia 57:660–671. https://doi.org/10.1007/s00125-014-3171-6
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (2010) Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 6:e1001233. https://doi.org/10.1371/journal.pgen.1001233
Cech T (1990) Self-splicing of group I introns. Annu Rev Biochem 59:543–568. https://doi.org/10.1146/annurev.bi.59.070190.002551
Chen J, Zou Q, Lv D, Wei Y, Raza MA, Chen Y, Li P, ** X, Xu H, Wen A, Zhu L, Tang G, Li M, Jiang A, Liu Y, Fu Y, Jiang Y, Li X (2018) Comprehensive transcriptional landscape of porcine cardiac and skeletal muscles reveals differences of aging. Oncotarget 9:1524–1541. https://doi.org/10.18632/oncotarget.23290
Chen LH, Chiou GY, Chen YW, Li HY, Chiou SH (2010) MicroRNA and aging: a novel modulator in regulating the aging network. Ageing Res Rev 9(Suppl 1):S59–S66. https://doi.org/10.1016/j.arr.2010.08.002
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17:205–211. https://doi.org/10.1038/nrm.2015.32
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388. https://doi.org/10.1080/15476286.2015.1020271
Chen X, Han P, Zhou T, Guo X, Song X, Li Y (2016) circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep 6:34985. https://doi.org/10.1038/srep34985
Cheng X, Joe B (2017) Circular RNAs in rat models of cardiovascular and renal diseases. Physiol Genomics 49:484–490. https://doi.org/10.1152/physiolgenomics.00064.2017
Cirrik S, S-S GW (2018) IGF-1 receptor cleavage in hypertension. Hypertens Res 41:406–413. https://doi.org/10.1038/s41440-018-0023-7
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134. https://doi.org/10.1016/j.cell.2015.02.014
Dang R, Liu F, Li Y (2017) Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys Res Commun 490:104–110. https://doi.org/10.1016/j.bbrc.2017.05.164
Dang RY, Liu FL, Li Y (2017) Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem Biophys Res Commun 490:104–110. https://doi.org/10.1016/j.bbrc.2017.05.164
Donlon TA, Curb JD, He Q, Grove JS, Masaki KH, Rodriguez B, Elliott A, Willcox DC, Willcox BJ (2012) FOXO3 gene variants and human aging: coding variants may not be key players. J Gerontol A Biol Sci Med Sci 67:1132–1139. https://doi.org/10.1093/gerona/gls067
Du W, Yang W, Chen Y, Wu Z, Foster F, Yang Z, Li X, Yang B (2017) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38:1402–1412. https://doi.org/10.1093/eurheartj/ehw001
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB (2017) Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 24:357–370. https://doi.org/10.1038/cdd.2016.133
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44:2846–2858. https://doi.org/10.1093/nar/gkw027
Dudekula D, Panda A, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13:34–42. https://doi.org/10.1080/15476286.2015.1128065
Eger N, Schoppe L, Schuster S, Laufs U, Boeckel J (2018) Circular RNA splicing. Adv Exp Med Biol 1087:41–52. https://doi.org/10.1007/978-981-13-1426-1_4
Gallego-Colon E, Sampson RD, Sattler S, Schneider MD, Rosenthal N, Tonkin J (2015) Cardiac-restricted IGF-1Ea overexpression reduces the early accumulation of inflammatory myeloid cells and mediates expression of extracellular matrix remodelling genes after myocardial infarction. Mediat Inflamm 2015:484357–484310. https://doi.org/10.1155/2015/484357
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J (2013) Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 4:283. https://doi.org/10.3389/fgene.2013.00283
Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. Rna 20:1666–1670. https://doi.org/10.1261/rna.043687.113
Gupta SK, Piccoli MT, Thum T (2014) Non-coding RNAs in cardiovascular ageing. Ageing Res Rev 17:79–85. https://doi.org/10.1016/j.arr.2014.01.002
Hasdai D, Rizza R, Holmes D, Richardson D, Cohen P, Lerman A (1998) Insulin and insulin-like growth factor-I cause coronary vasorelaxation in vitro. Hypertension 32:228–234. https://doi.org/10.1161/01.hyp.32.2.228
Higashi Y, Gautam S, Delafontaine P, Sukhanov S (2019) IGF-1 and cardiovascular disease. Growth Hormon IGF Res 45:6–16. https://doi.org/10.1016/j.ghir.2019.01.002
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7:12429. https://doi.org/10.1038/ncomms12429
Huang JL, Qin MC, Zhou Y, Xu ZH, Yang SM, Zhang F, Zhong J, Liang MK, Chen B, Zhang WY, Wu DP, Zhong ZG (2018) Comprehensive analysis of differentially expressed profiles of Alzheimer’s disease associated circular RNAs in an Alzheimer’s disease mouse model. Aging (Albany NY) 10:253–265. https://doi.org/10.18632/aging.101387
Huang X, Chen Y, **ao J, Huang Z, He L, Xu D, Peng J (2018) Identification of differentially expressed circular RNAs during TGF-ss1-induced endothelial-to-mesenchymal transition in rat coronary artery endothelial cells. Anatol J Cardiol 19:192–197. https://doi.org/10.14744/AnatolJCardiol.2018.95142
Janga S, Mittal N (2011) Construction, structure and dynamics of post-transcriptional regulatory network directed by RNA-binding proteins. Adv Exp Med Biol 722:103–117. https://doi.org/10.1007/978-1-4614-0332-6_7
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. https://doi.org/10.1038/nbt.2890
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 19:141–157. https://doi.org/10.1261/rna.035667.112
Jiang J, Yang Y, Jiang R (2016) Regulating mechanisms of circRNA and their relationship with cardiovascular diseases. Zhonghua **n Xue Guan Bing Za Zhi 44:364–366. https://doi.org/10.3760/cma.j.issn.0253-3758.2016.04.021
Joseph P, Leong D, McKee M, Anand SS, Schwalm JD, Teo K, Mente A, Yusuf S (2017) Reducing the global burden of cardiovascular disease, part 1: the epidemiology and risk factors. Circ Res 121:677–694. https://doi.org/10.1161/circresaha.117.308903
Jost I, Shalamova LA, Gerresheim GK, Niepmann M, Bindereif A, Rossbach O (2018) Functional sequestration of microRNA-122 from hepatitis C virus by circular RNA sponges. RNA Biol 15:1032–1039. https://doi.org/10.1080/15476286.2018.1435248
Jufri NF, Mohamedali A, Avolio A, Baker MS (2015) Mechanical stretch: physiological and pathological implications for human vascular endothelial cells. Vasc Cell 7:8. https://doi.org/10.1186/s13221-015-0033-z
Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29:2168–2182. https://doi.org/10.1101/gad.270421.115
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. Rna 20:1829–1842. https://doi.org/10.1261/rna.047126.114
Lei W, Feng T, Fang X, Yu Y, Yang J, Zhao ZA, Liu J, Shen Z, Deng W, Hu S (2018) Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther 9:56. https://doi.org/10.1186/s13287-018-0793-5
Li CY, Ma L, Yu B (2017) Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed Pharmacother 95:1514–1519. https://doi.org/10.1016/j.biopha.2017.09.064
Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y (2015) Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 6:6001–6013. https://doi.org/10.18632/oncotarget.3469
Li H, Wei X, Yang J, Dong D, Hao D, Huang Y, Lan X, Plath M, Lei C, Ma Y, Lin F, Bai Y, Chen H (2018) circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol Ther Nucleic Acids 11:272–283. https://doi.org/10.1016/j.omtn.2018.02.012
Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, Zheng Q, Li Y, Wang P, He X, Huang S (2018) exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res 46:D106–d112. https://doi.org/10.1093/nar/gkx891
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264. https://doi.org/10.1038/nsmb.2959
Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28:2233–2247. https://doi.org/10.1101/gad.251926.114
Lin H, Yue Y, Maidana DE, Bouzika P, Atik A, Matsumoto H, Miller JW, Vavvas DG (2016) Drug delivery nanoparticles: toxicity comparison in retinal pigment epithelium and retinal vascular endothelial cells. Semin Ophthalmol 31:1–9. https://doi.org/10.3109/08820538.2015.1114865
Liu M, Wang Q, Shen J, Yang B, Ding X (2019) Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol 16:899–905. https://doi.org/10.1080/15476286.2019.1600395
Liu W, Ma W, Yuan Y, Zhang Y, Sun S (2018) Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun 500:846–851. https://doi.org/10.1016/j.bbrc.2018.04.172
Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, Liu CC, Huang HD (2016) CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 44:D209–D215. https://doi.org/10.1093/nar/gkv940
Mattick J (2001) Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep 2:986–991. https://doi.org/10.1093/embo-reports/kve230
Mehta S, Dempsey R, Vemuganti R (2020) Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol 186:101746. https://doi.org/10.1016/j.pneurobio.2020.101746
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Molina-Sánchez M, Martinez-Abarca F, Toro N (2006) Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J Biol Chem 281:28737–28744. https://doi.org/10.1074/jbc.M602695200
Pan RY, Liu P, Zhou HT, Sun WX, Song J, Shu J, Cui GJ, Yang ZJ, Jia EZ (2017) Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. Oncotarget 8:60280–60290. https://doi.org/10.18632/oncotarget.19941
Pyle A (2016) Group II intron self-splicing. Annu Rev Biophys 45:183–205. https://doi.org/10.1146/annurev-biophys-062215-011149
Rodríguez-Trelles F, Tarrío R, Ayala F (2006) Origins and evolution of spliceosomal introns. Annu Rev Genet 40:47–76. https://doi.org/10.1146/annurev.genet.40.110405.090625
Rodriguez BV, Malczewskyj ET, Cabiya JM, Lewis LK, Maeder C (2016) Identification of RNase-resistant RNAs in Saccharomyces cerevisiae extracts: Separation from chromosomal DNA by selective precipitation. Anal Biochem 492:69–75. https://doi.org/10.1016/j.ab.2015.09.017
Salgado-Somoza A, Zhang L, Vausort M, Devaux Y (2017) The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc 17:33–36. https://doi.org/10.1016/j.ijcha.2017.11.001
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. https://doi.org/10.1371/journal.pone.0030733
Sang M, Meng L, Sang Y, Liu S, Ding P, Ju Y, Liu F, Gu L, Lian Y, Li J, Wu Y, Zhang X, Shan B (2018) Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett 426:37–46. https://doi.org/10.1016/j.canlet.2018.03.049
Shang FF, Luo S, Liang X, **a Y (2018) Alterations of circular RNAs in hyperglycemic human endothelial cells. Biochem Biophys Res Commun 499:551–555. https://doi.org/10.1016/j.bbrc.2018.03.187
Sun M, An Q, Chen L, Guo L (2020) MIR-520f regulated itch expression and promoted cell proliferation in human melanoma cells. Dose-Response 18:1559325820918450. https://doi.org/10.1177/1559325820918450
Sun Y, Yang Z, Zheng B, Zhang XH, Zhang ML, Zhao XS, Zhao HY, Suzuki T, Wen JK (2017) A novel regulatory mechanism of smooth muscle alpha-actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ Res 121:628–635. https://doi.org/10.1161/circresaha.117.311441
Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD, Li PY, Richards AM, Foo RS (2017) A landscape of circular RNA expression in the human heart. Cardiovasc Res 113:298–309. https://doi.org/10.1093/cvr/cvw250
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, **ao Z, Zhang Z, Lin QX, Zheng XL, Yang M, Wu SL, Cheng JD, Shan ZX (2017) CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 7:40342. https://doi.org/10.1038/srep40342
Tatomer D, Liang D, Wilusz J (2017) Inducible expression of eukaryotic circular RNAs from plasmids. Methods Mol Biol 1648:143–154. https://doi.org/10.1007/978-1-4939-7204-3_11
Toptan T, Abere B, Nalesnik M, Swerdlow S, Ranganathan S, Lee N, Shair K, Moore P, Chang Y (2018) Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A 115:E8737–E8745. https://doi.org/10.1073/pnas.1811728115
Wang K, Gan T, Li N, Liu C, Zhou L, Gao J, Chen C, Yan K, Ponnusamy M, Zhang Y, Li P (2017) Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 24:1111–1120. https://doi.org/10.1038/cdd.2017.61
Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37:2602–2611. https://doi.org/10.1093/eurheartj/ehv713
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J (2014) Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9:e90859. https://doi.org/10.1371/journal.pone.0090859
Wang YS, Zhou J, Hong K, Cheng XS, Li YG (2015) MicroRNA-223 displays a protective role against cardiomyocyte hypertrophy by targeting cardiac troponin I-interacting kinase. Cell Physiol Biochem 35:1546–1556. https://doi.org/10.1159/000373970
Weber C, Noels H (2011) Atherosclerosis: current pathogenesis and therapeutic options. Nat Med 17:1410–1422. https://doi.org/10.1038/nm.2538
Wilusz JE, Sharp PA (2013) Molecular biology. A circuitous route to noncoding RNA. Science 340:440–441. https://doi.org/10.1126/science.1238522
Wu N, ** L, Cai J (2017) Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin Exp Hypertens 39:454–459. https://doi.org/10.1080/10641963.2016.1273944
**a S, Feng J, Chen K, Ma Y, Gong J, Cai F, ** Y, Gao Y, **a L, Chang H, Wei L, Han L, He C (2018) CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res 46:D925–d929. https://doi.org/10.1093/nar/gkx863
**e YZ, Yang F, Tan W, Li X, Jiao C, Huang R, Yang BB (2016) The anti-cancer components of Ganoderma lucidum possesses cardiovascular protective effect by regulating circular RNA expression. Oncoscience 3:203–207. https://doi.org/10.18632/oncoscience.316
Ye S, Willeit J, Kronenberg F, Xu Q, Kiechl S (2008) Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J Am Coll Cardiol 52:378–384. https://doi.org/10.1016/j.jacc.2007.11.087
Zang J, Lu D, Xu A (2020) The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res 98:87–97. https://doi.org/10.1002/jnr.24356
Zhang X, Qiu S, Luo P, Zhou H, **g W, Liang C, Tu J (2018) Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark 22:135–142. https://doi.org/10.3233/cbm-171109
Zhang Y, Yang L, Chen LL (2014) Life without A tail: new formats of long noncoding RNAs. Int J Biochem Cell Biol 54:338–349. https://doi.org/10.1016/j.biocel.2013.10.009
Zhang Y, Zhang XO, Chen T, **ang JF, Yin QF, **ng YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806. https://doi.org/10.1016/j.molcel.2013.08.017
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M (2017) Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 7:39918. https://doi.org/10.1038/srep39918
Zhao Z, Wang K, Wu F, Wang W, Zhang K, Hu H, Liu Y, Jiang T (2018) circRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis 9:475. https://doi.org/10.1038/s41419-018-0503-3
Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL, Zhou H, Yang JH, Qu LH (2016) deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res 44:D196–D202. https://doi.org/10.1093/nar/gkv1273
Zhou B, Yu JW (2017) A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun 487:769–775. https://doi.org/10.1016/j.bbrc.2017.04.044
Funding
This work was financially supported by Young Elite Scientists Sponsorship Program by CAST (No. CACM-2018-QNRC2-B04) and Young Prominant Medical Professionals Program by Tian**.
Author information
Authors and Affiliations
Contributions
Yuejia Ding and Chunmiao Lu wrote the paper. Wanqin Zhang, Yuan Wang, and Yanyang Li made the literature selection. Shichao Lv, Ya** Zhu, and Jun** Zhang revised the paper.
The authors declare that all data were generated in-house and that no paper mill was used.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Informed consent
Not applicable
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key points
• CircRNAs exhibit important functions in cardiovascular disease.
• The biogenesis of circRNAs can be divided into two ways
• CircRNAs may become therapeutic targets for cardiovascular diseases
Rights and permissions
About this article
Cite this article
Ding, Y., Lu, C., Zhang, W. et al. The emerging role of circular RNAs in cardiovascular diseases. J Physiol Biochem 77, 343–353 (2021). https://doi.org/10.1007/s13105-021-00807-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13105-021-00807-y